You are browsing environment: FUNGIDB
CAZyme Information: EJT71317.1
You are here: Home > Sequence: EJT71317.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gaeumannomyces tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici | |||||||||||
| CAZyme ID | EJT71317.1 | |||||||||||
| CAZy Family | CBM21 | |||||||||||
| CAZyme Description | Feruloyl esterase C [Source:UniProtKB/TrEMBL;Acc:J3PAQ1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 104 | 236 | 6e-20 | 0.5859030837004405 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 1.37e-19 | 100 | 332 | 39 | 290 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 224423 | DAP2 | 1.17e-07 | 102 | 235 | 372 | 506 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| 226584 | COG4099 | 0.005 | 110 | 236 | 176 | 303 | Predicted peptidase [General function prediction only]. |
| 223489 | DLH | 0.007 | 151 | 258 | 62 | 169 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.13e-114 | 77 | 335 | 88 | 346 | |
| 3.75e-113 | 87 | 332 | 47 | 292 | |
| 5.31e-113 | 87 | 332 | 47 | 292 | |
| 1.33e-112 | 80 | 332 | 39 | 291 | |
| 4.55e-112 | 81 | 332 | 37 | 288 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.89e-77 | 77 | 317 | 18 | 249 | Probable feruloyl esterase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=faeC PE=3 SV=1 |
|
| 2.35e-76 | 77 | 317 | 18 | 249 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-1 PE=3 SV=1 |
|
| 4.70e-76 | 77 | 317 | 18 | 249 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=faeC PE=3 SV=1 |
|
| 4.70e-76 | 77 | 317 | 18 | 249 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=faeC PE=3 SV=1 |
|
| 2.49e-75 | 81 | 317 | 20 | 247 | Probable feruloyl esterase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
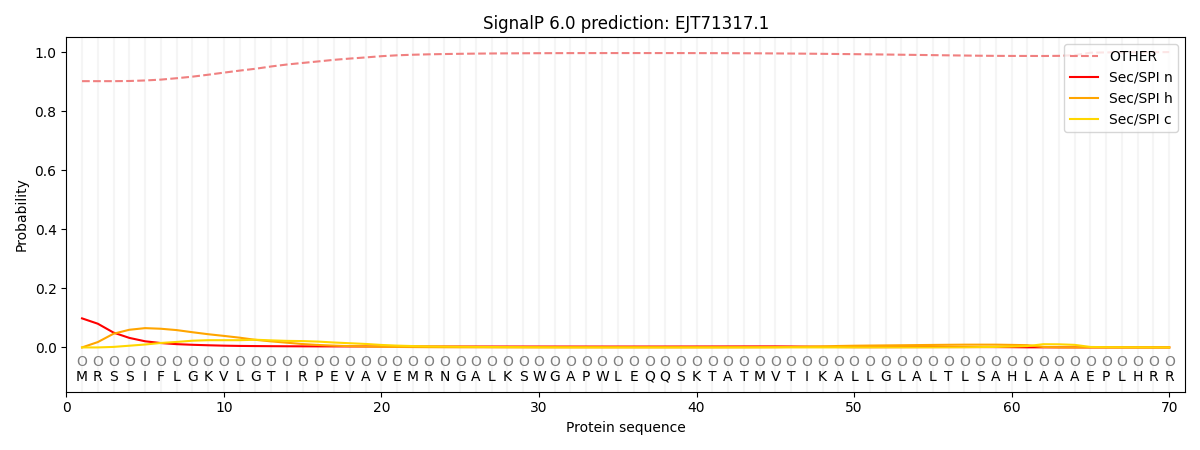
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.914952 | 0.085039 |
