You are browsing environment: FUNGIDB
CAZyme Information: EJT71307.1
You are here: Home > Sequence: EJT71307.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gaeumannomyces tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici | |||||||||||
| CAZyme ID | EJT71307.1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | Cutinase [Source:UniProtKB/TrEMBL;Acc:J3PAP1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.74:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 40 | 208 | 3.2e-47 | 0.9841269841269841 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 2.93e-59 | 40 | 208 | 2 | 172 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.18e-68 | 1 | 204 | 1 | 199 | |
| 1.18e-68 | 1 | 204 | 1 | 199 | |
| 5.80e-68 | 38 | 205 | 38 | 206 | |
| 3.42e-67 | 32 | 209 | 23 | 200 | |
| 6.87e-67 | 40 | 205 | 31 | 196 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.09e-52 | 40 | 205 | 79 | 243 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 2.76e-27 | 36 | 195 | 19 | 184 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 3.05e-26 | 40 | 205 | 31 | 203 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 3.05e-26 | 40 | 205 | 31 | 203 | Chain A, CUTINASE [Fusarium vanettenii],1CUW_B Chain B, CUTINASE [Fusarium vanettenii] |
|
| 3.05e-26 | 40 | 205 | 31 | 203 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.22e-67 | 40 | 205 | 31 | 196 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 4.48e-65 | 40 | 205 | 31 | 195 | Cutinase OS=Monilinia fructicola OX=38448 GN=CUT1 PE=2 SV=1 |
|
| 1.53e-53 | 32 | 209 | 24 | 201 | Cutinase pbc1 OS=Pyrenopeziza brassicae OX=76659 GN=pbc1 PE=1 SV=1 |
|
| 4.71e-52 | 40 | 205 | 79 | 243 | Cutinase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_60489 PE=3 SV=2 |
|
| 4.71e-52 | 40 | 205 | 79 | 243 | Cutinase OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=M419DRAFT_76732 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
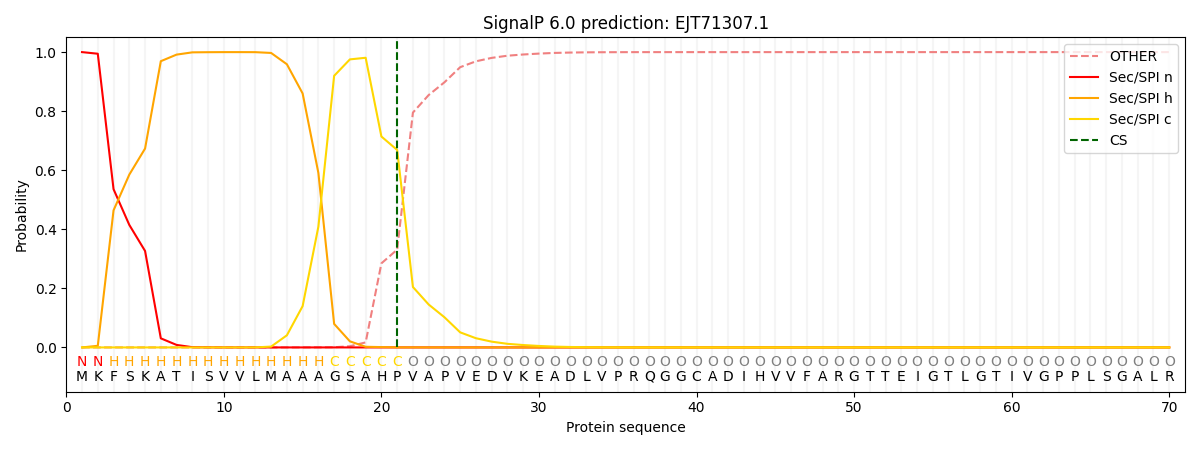
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000436 | 0.999521 | CS pos: 21-22. Pr: 0.6686 |
