You are browsing environment: FUNGIDB
EJT68770.1
Basic Information
help
Species
Gaeumannomyces tritici
Lineage
Ascomycota; Sordariomycetes; ; Magnaporthaceae; Gaeumannomyces; Gaeumannomyces tritici
CAZyme ID
EJT68770.1
CAZy Family
AA11
CAZyme Description
unspecified product
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
522
GL385423|CGC1 57023.09
9.6970
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_GtriticiR3-111a-1
14749
644352
291
14458
Gene Location
No EC number prediction in EJT68770.1.
Family
Start
End
Evalue
family coverage
GH28
171
498
9e-48
0.9323076923076923
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
227721
Pgu1
1.75e-22
146
521
82
517
Polygalacturonase [Carbohydrate transport and metabolism].
more
403800
Pectate_lyase_3
7.26e-07
146
243
1
103
Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28.
more
395231
Glyco_hydro_28
0.004
184
433
12
243
Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
0.0
1
521
1
521
8.06e-231
27
521
22
513
2.17e-228
23
521
18
513
8.28e-198
7
521
4
518
8.37e-158
19
509
14
503
EJT68770.1 has no PDB hit.
EJT68770.1 has no Swissprot hit.
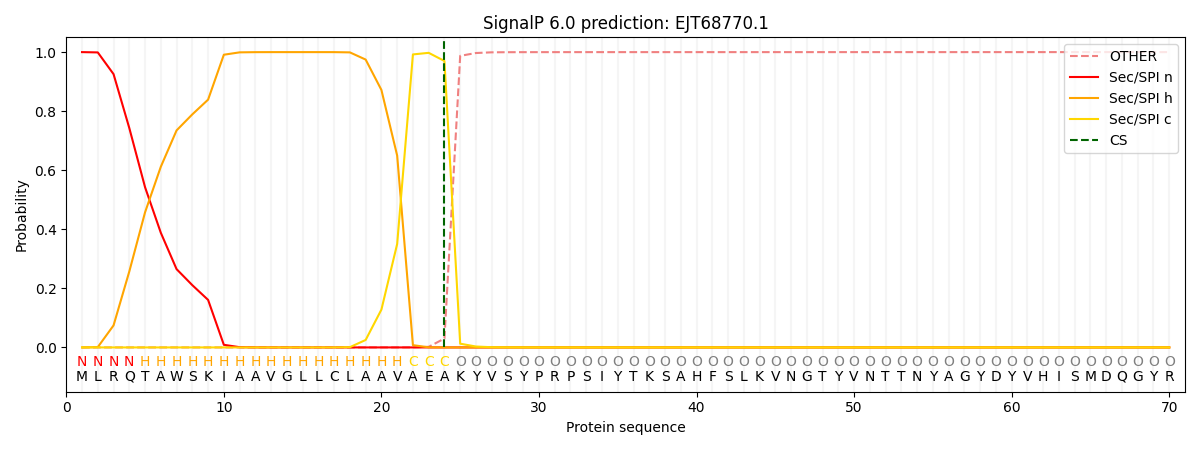
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000227
0.999727
CS pos: 24-25. Pr: 0.9699
There is no transmembrane helices in EJT68770.1.