You are browsing environment: FUNGIDB
CAZyme Information: EJT51018.1
You are here: Home > Sequence: EJT51018.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichosporon asahii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Trichosporonaceae; Trichosporon; Trichosporon asahii | |||||||||||
| CAZyme ID | EJT51018.1 | |||||||||||
| CAZy Family | GT24 | |||||||||||
| CAZyme Description | Cellulase domain-containing protein [Source:UniProtKB/TrEMBL;Acc:J5R710] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:29 | 3.2.1.21:6 | 3.2.1.75:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 381 | 635 | 6.4e-58 | 0.7953795379537953 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 3.74e-22 | 332 | 551 | 16 | 227 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 5.36e-07 | 385 | 629 | 23 | 245 | Cellulase (glycosyl hydrolase family 5). |
| 165513 | PHA03255 | 0.003 | 252 | 327 | 56 | 124 | BDLF3; Provisional |
| 381626 | GH113_mannanase-like | 0.003 | 378 | 447 | 11 | 81 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
| 236776 | PRK10856 | 0.007 | 249 | 314 | 164 | 229 | cytoskeleton protein RodZ. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.81e-46 | 321 | 623 | 72 | 363 | |
| 1.81e-46 | 321 | 623 | 72 | 363 | |
| 3.21e-45 | 321 | 623 | 72 | 363 | |
| 1.55e-44 | 321 | 616 | 69 | 355 | |
| 2.13e-44 | 321 | 616 | 72 | 356 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.54e-35 | 334 | 582 | 9 | 252 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 1.40e-34 | 334 | 582 | 9 | 252 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 1.53e-34 | 334 | 582 | 14 | 257 | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution [Candida albicans SC5314],4M81_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution [Candida albicans SC5314],4M82_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution [Candida albicans SC5314] |
|
| 1.56e-34 | 334 | 582 | 15 | 258 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
|
| 1.56e-34 | 334 | 582 | 15 | 258 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.79e-37 | 334 | 605 | 42 | 296 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=exgA PE=3 SV=2 |
|
| 5.36e-37 | 332 | 650 | 37 | 346 | Glucan 1,3-beta-glucosidase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=exg1 PE=2 SV=1 |
|
| 3.00e-36 | 306 | 602 | 19 | 293 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
|
| 3.59e-36 | 333 | 605 | 36 | 296 | Glucan 1,3-beta-glucosidase OS=Blumeria graminis OX=34373 PE=3 SV=1 |
|
| 2.93e-35 | 332 | 620 | 27 | 297 | Probable glucan 1,3-beta-glucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=exgA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
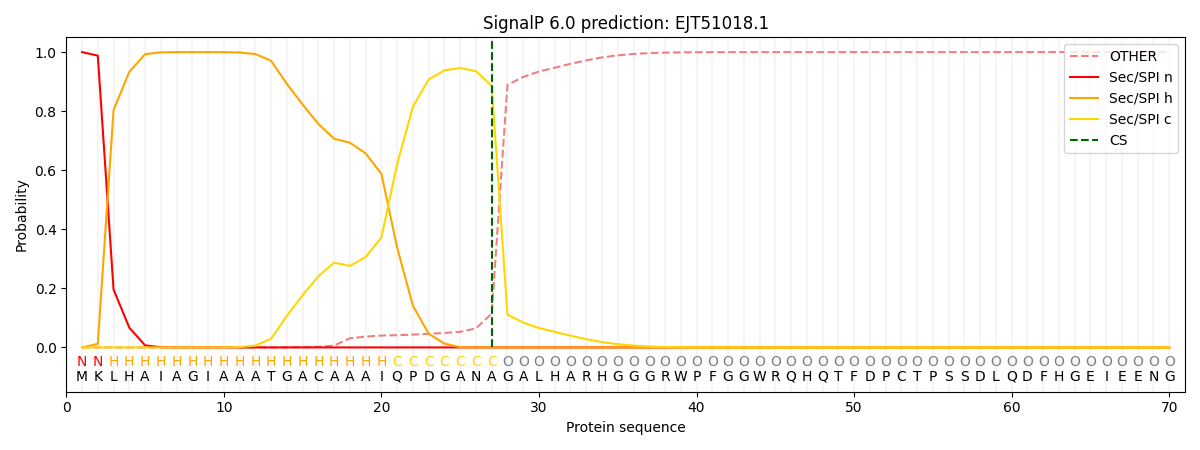
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000511 | 0.999440 | CS pos: 27-28. Pr: 0.8855 |
