You are browsing environment: FUNGIDB
CAZyme Information: EHY58286.1
You are here: Home > Sequence: EHY58286.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
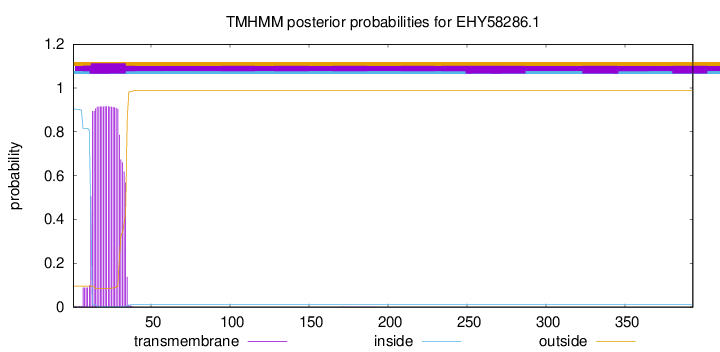
TMHMM annotations
Basic Information help
| Species | Exophiala dermatitidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala dermatitidis | |||||||||||
| CAZyme ID | EHY58286.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 153 | 271 | 5.1e-16 | 0.44715447154471544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173412 | PTZ00121 | 1.21e-14 | 295 | 389 | 1345 | 1439 | MAEBL; Provisional |
| 173412 | PTZ00121 | 1.46e-12 | 308 | 389 | 1397 | 1478 | MAEBL; Provisional |
| 368535 | Glyco_transf_34 | 3.67e-12 | 81 | 267 | 40 | 231 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 173412 | PTZ00121 | 1.21e-11 | 310 | 389 | 1412 | 1491 | MAEBL; Provisional |
| 173412 | PTZ00121 | 2.13e-11 | 281 | 389 | 1357 | 1465 | MAEBL; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.67e-71 | 54 | 313 | 207 | 454 | |
| 3.96e-70 | 45 | 307 | 165 | 415 | |
| 7.83e-69 | 54 | 311 | 144 | 387 | |
| 7.83e-69 | 54 | 311 | 144 | 387 | |
| 7.83e-69 | 54 | 311 | 144 | 387 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
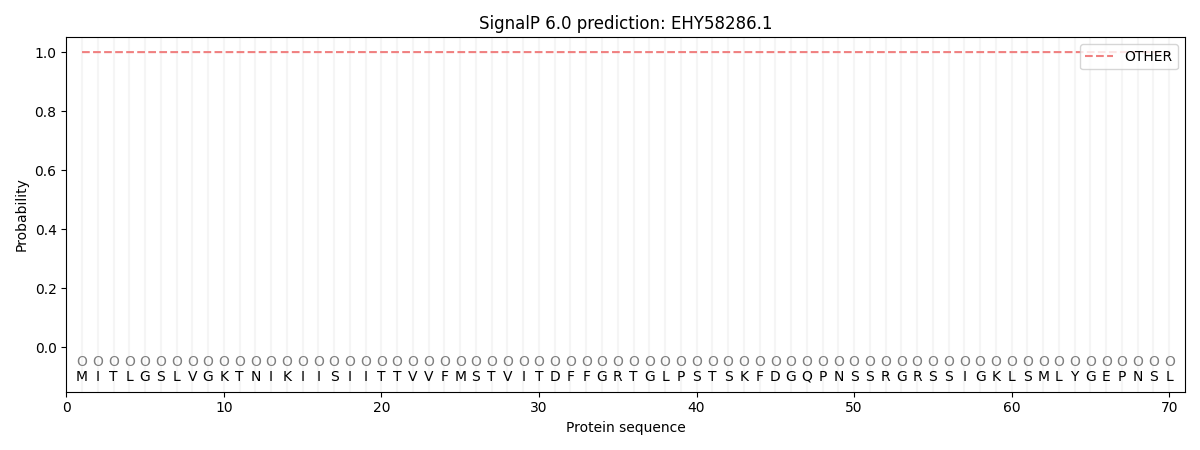
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999966 | 0.000057 |