You are browsing environment: FUNGIDB
CAZyme Information: EHY56479.1
You are here: Home > Sequence: EHY56479.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala dermatitidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala dermatitidis | |||||||||||
| CAZyme ID | EHY56479.1 | |||||||||||
| CAZy Family | GH32 | |||||||||||
| CAZyme Description | beta-glucosidase, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.37:8 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 68 | 318 | 3.9e-61 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178629 | PLN03080 | 1.81e-176 | 35 | 754 | 43 | 768 | Probable beta-xylosidase; Provisional |
| 185053 | PRK15098 | 1.43e-52 | 102 | 766 | 116 | 764 | beta-glucosidase BglX. |
| 224389 | BglX | 8.11e-45 | 101 | 419 | 78 | 365 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| 396478 | Glyco_hydro_3_C | 9.79e-39 | 390 | 617 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| 395747 | Glyco_hydro_3 | 1.35e-32 | 100 | 350 | 82 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 2 | 783 | 3 | 773 | |
| 0.0 | 21 | 780 | 25 | 772 | |
| 0.0 | 10 | 780 | 9 | 766 | |
| 0.0 | 12 | 780 | 12 | 780 | |
| 0.0 | 21 | 780 | 25 | 763 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.05e-280 | 21 | 779 | 2 | 742 | Chain A, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
|
| 3.05e-280 | 21 | 779 | 2 | 742 | Chain A, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
|
| 4.44e-228 | 21 | 781 | 29 | 766 | GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
|
| 3.47e-225 | 21 | 777 | 29 | 764 | Chain A, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
|
| 3.59e-225 | 21 | 777 | 29 | 764 | Chain A, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.90e-292 | 14 | 780 | 20 | 768 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=bxlB PE=3 SV=1 |
|
| 7.90e-292 | 14 | 780 | 20 | 768 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bxlB PE=3 SV=1 |
|
| 2.41e-288 | 8 | 780 | 16 | 768 | Probable exo-1,4-beta-xylosidase xlnD OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xlnD PE=3 SV=1 |
|
| 4.83e-284 | 17 | 780 | 25 | 759 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=bxlB PE=3 SV=1 |
|
| 1.88e-281 | 5 | 779 | 13 | 763 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bxlB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
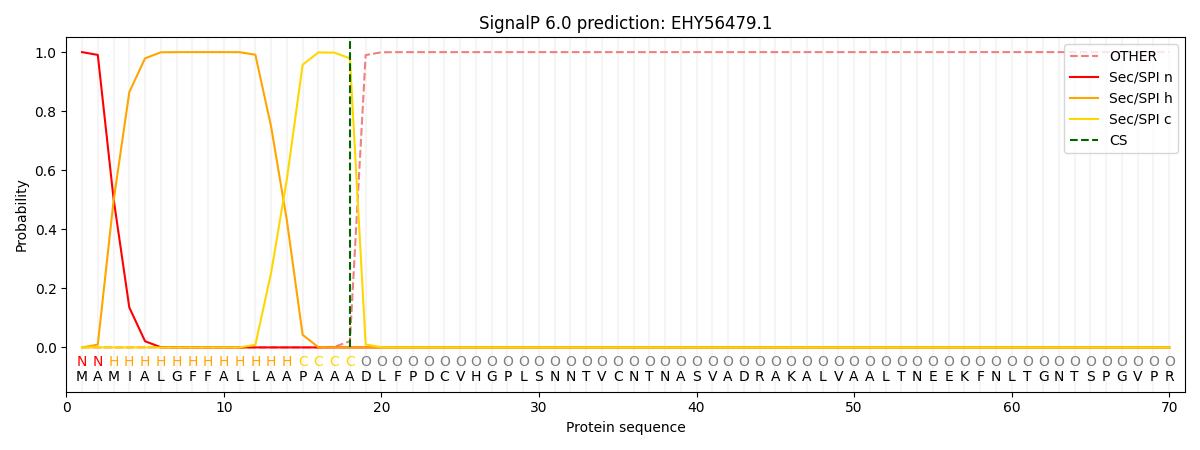
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000230 | 0.999718 | CS pos: 18-19. Pr: 0.9791 |
