You are browsing environment: FUNGIDB
CAZyme Information: EHY54535.1
You are here: Home > Sequence: EHY54535.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala dermatitidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala dermatitidis | |||||||||||
| CAZyme ID | EHY54535.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | extracellular cell wall glucanase Crf1/allergen Asp F9 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 73 | 234 | 8.6e-60 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185692 | GH16_fungal_CRH1_transglycosylase | 1.17e-86 | 48 | 253 | 2 | 200 | glycosylphosphatidylinositol-glucanosyltransferase. Group of fungal GH16 members related to Saccharomyces cerevisiae Crh1p. Chr1p and Crh2p are transglycosylases that are required for the linkage of chitin to beta(1-3)glucose branches of beta(1-6)glucan, an important step in the assembly of new cell wall. Both have been shown to be glycosylphosphatidylinositol (GPI)-anchored. A third homologous protein, Crr1p, functions in the formation of the spore wall. They belongs to the family 16 of glycosyl hydrolases that includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| 395585 | Glyco_hydro_16 | 2.35e-46 | 73 | 231 | 11 | 166 | Glycosyl hydrolases family 16. |
| 240257 | PTZ00073 | 6.59e-42 | 691 | 761 | 12 | 82 | 60S ribosomal protein L37; Provisional |
| 396473 | Ribosomal_L37e | 5.66e-25 | 691 | 733 | 11 | 53 | Ribosomal protein L37e. This family includes ribosomal protein L37 from eukaryotes and archaebacteria. The family contains many conserved cysteines and histidines suggesting that this protein may bind to zinc. |
| 185683 | Glyco_hydrolase_16 | 3.43e-24 | 48 | 253 | 2 | 210 | glycosyl hydrolase family 16. The O-Glycosyl hydrolases are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycosyl hydrolase family 16. Family 16 includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.14e-73 | 27 | 296 | 29 | 299 | |
| 5.14e-73 | 27 | 296 | 29 | 299 | |
| 5.14e-73 | 27 | 296 | 29 | 299 | |
| 5.14e-72 | 27 | 296 | 29 | 299 | |
| 2.75e-69 | 7 | 280 | 2 | 269 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.15e-70 | 27 | 274 | 4 | 242 | Apo Crh5 transglycosylase [Aspergillus fumigatus Af293],6IBU_B Apo Crh5 transglycosylase [Aspergillus fumigatus Af293],6IBW_A Crh5 transglycosylase in complex with NAG [Aspergillus fumigatus Af293],6IBW_B Crh5 transglycosylase in complex with NAG [Aspergillus fumigatus Af293] |
|
| 7.61e-39 | 691 | 771 | 12 | 92 | Chain l1, Ribosomal protein L37 [Neurospora crassa] |
|
| 2.92e-38 | 691 | 768 | 12 | 89 | Chain Lj, Ribosomal protein L37 [Thermochaetoides thermophila DSM 1495],7OLD_Lj Chain Lj, Ribosomal protein L37 [Thermochaetoides thermophila DSM 1495] |
|
| 1.80e-31 | 691 | 763 | 11 | 83 | Chain j, eL37 [Saccharomyces cerevisiae] |
|
| 5.58e-31 | 691 | 761 | 11 | 81 | Chain AF, 60S ribosomal protein L37-A [Saccharomyces cerevisiae S288C],6TNU_AF Chain AF, 60S ribosomal protein L37-A [Saccharomyces cerevisiae],7B7D_Lf Chain Lf, 60S ribosomal protein L37-A [Saccharomyces cerevisiae S288C],7NRC_Ll Chain Ll, 60S ribosomal protein L37-A [Saccharomyces cerevisiae S288C],7NRD_Ll Chain Ll, 60S ribosomal protein L37-A [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.04e-68 | 16 | 274 | 11 | 263 | Probable glycosidase crf1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=crf1 PE=1 SV=2 |
|
| 1.06e-58 | 11 | 274 | 7 | 256 | Probable extracellular glycosidase ARB_05253 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_05253 PE=1 SV=2 |
|
| 3.19e-55 | 27 | 274 | 40 | 275 | Probable glycosidase CRH1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CRH1 PE=1 SV=1 |
|
| 2.65e-51 | 22 | 274 | 19 | 260 | Extracellular glycosidase CRH11 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CRH11 PE=1 SV=1 |
|
| 2.46e-48 | 27 | 274 | 31 | 269 | Probable extracellular glycosidase ARB_03382 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_03382 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
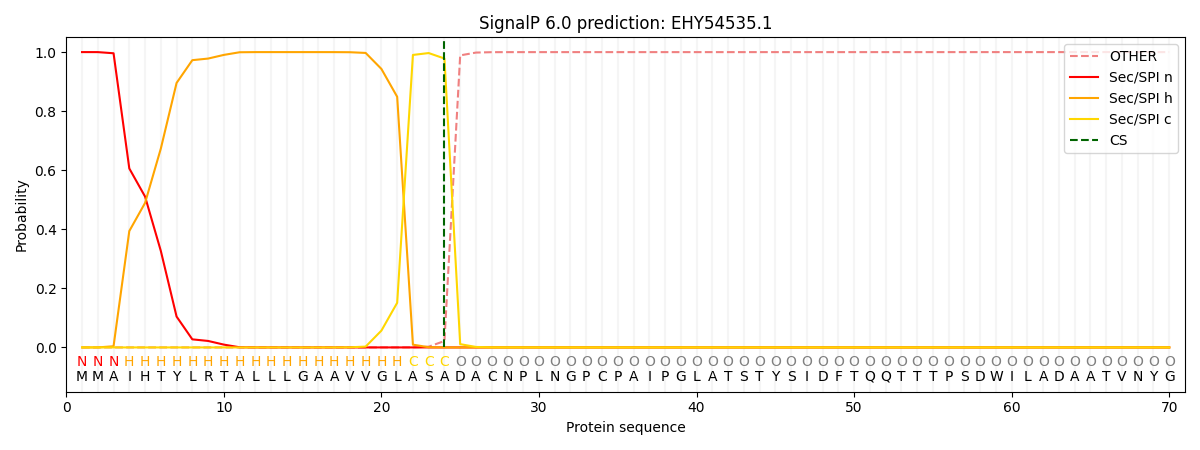
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000205 | 0.999770 | CS pos: 24-25. Pr: 0.9783 |
