You are browsing environment: FUNGIDB
CAZyme Information: EGD90016.1
You are here: Home > Sequence: EGD90016.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
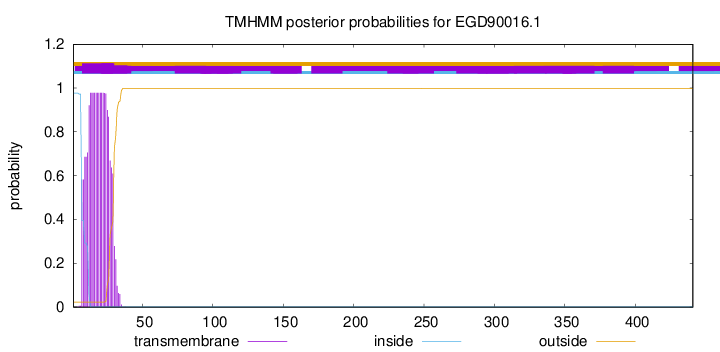
TMHMM annotations
Basic Information help
| Species | Trichophyton rubrum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Arthrodermataceae; Trichophyton; Trichophyton rubrum | |||||||||||
| CAZyme ID | EGD90016.1 | |||||||||||
| CAZy Family | GH63 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT32 | 133 | 212 | 5.8e-18 | 0.9111111111111111 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226297 | OCH1 | 4.66e-21 | 112 | 242 | 82 | 208 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 398274 | Gly_transf_sug | 1.40e-14 | 134 | 213 | 9 | 90 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 173412 | PTZ00121 | 8.51e-05 | 364 | 439 | 1308 | 1392 | MAEBL; Provisional |
| 274303 | tolA_full | 0.001 | 364 | 434 | 119 | 189 | TolA protein. TolA couples the inner membrane complex of itself with TolQ and TolR to the outer membrane complex of TolB and OprL (also called Pal). Most of the length of the protein consists of low-complexity sequence that may differ in both length and composition from one species to another, complicating efforts to discriminate TolA (the most divergent gene in the tol-pal system) from paralogs such as TonB. Selection of members of the seed alignment and criteria for setting scoring cutoffs are based largely conserved operon struction. //The Tol-Pal complex is required for maintaining outer membrane integrity. Also involved in transport (uptake) of colicins and filamentous DNA, and implicated in pathogenesis. Transport is energized by the proton motive force. TolA is an inner membrane protein that interacts with periplasmic TolB and with outer membrane porins ompC, phoE and lamB. [Transport and binding proteins, Other, Cellular processes, Pathogenesis] |
| 173412 | PTZ00121 | 0.002 | 364 | 441 | 1496 | 1590 | MAEBL; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.31e-61 | 109 | 385 | 143 | 420 | |
| 9.29e-61 | 109 | 379 | 95 | 366 | |
| 2.26e-60 | 109 | 379 | 129 | 400 | |
| 9.21e-60 | 109 | 381 | 107 | 380 | |
| 1.85e-59 | 106 | 374 | 80 | 350 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.52e-21 | 112 | 364 | 142 | 394 | Initiation-specific alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=och1 PE=1 SV=2 |
|
| 1.28e-19 | 113 | 364 | 100 | 376 | Initiation-specific alpha-1,6-mannosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=OCH1 PE=3 SV=1 |
|
| 1.91e-19 | 113 | 365 | 131 | 389 | Putative glycosyltransferase HOC1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HOC1 PE=1 SV=3 |
|
| 4.78e-12 | 112 | 275 | 96 | 295 | Initiation-specific alpha-1,6-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=OCH1 PE=1 SV=1 |
|
| 7.57e-08 | 112 | 203 | 62 | 151 | Mannosyl phosphorylinositol ceramide synthase CSH1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CSH1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999966 | 0.000058 |