You are browsing environment: FUNGIDB
CAZyme Information: EFQ32679.1
You are here: Home > Sequence: EFQ32679.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
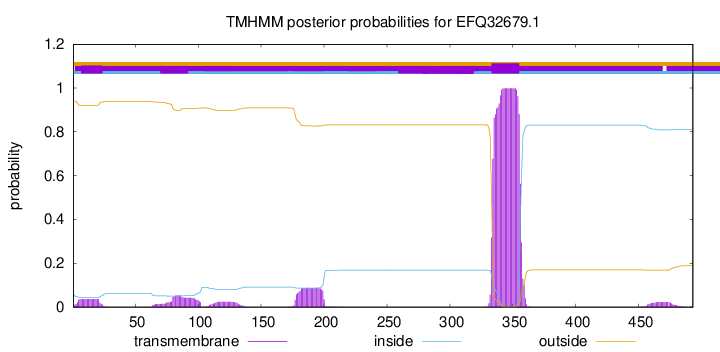
TMHMM annotations
Basic Information help
| Species | Colletotrichum graminicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Glomerellaceae; Colletotrichum; Colletotrichum graminicola | |||||||||||
| CAZyme ID | EFQ32679.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | GDSL-like Lipase/Acylhydrolase [Source:UniProtKB/TrEMBL;Acc:E3QP91] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE2 | 169 | 339 | 9.1e-17 | 0.6985645933014354 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238869 | Endoglucanase_E_like | 1.22e-26 | 169 | 466 | 1 | 169 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
| 238141 | SGNH_hydrolase | 2.50e-07 | 237 | 336 | 13 | 112 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 404371 | Lipase_GDSL_2 | 6.16e-05 | 290 | 336 | 62 | 106 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238865 | sialate_O-acetylesterase_like1 | 1.72e-04 | 287 | 335 | 67 | 113 | sialate O-acetylesterase_like family of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.55e-242 | 1 | 493 | 2 | 400 | |
| 1.27e-197 | 17 | 493 | 19 | 433 | |
| 2.19e-193 | 16 | 493 | 17 | 432 | |
| 3.59e-193 | 10 | 493 | 12 | 436 | |
| 1.45e-192 | 10 | 493 | 12 | 436 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.75e-08 | 47 | 337 | 5 | 260 | Chain A, ENDOGLUCANASE E [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.28e-08 | 40 | 337 | 479 | 741 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001544 | 0.998434 | CS pos: 18-19. Pr: 0.9000 |