You are browsing environment: FUNGIDB
CAZyme Information: EFQ31050.1
You are here: Home > Sequence: EFQ31050.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
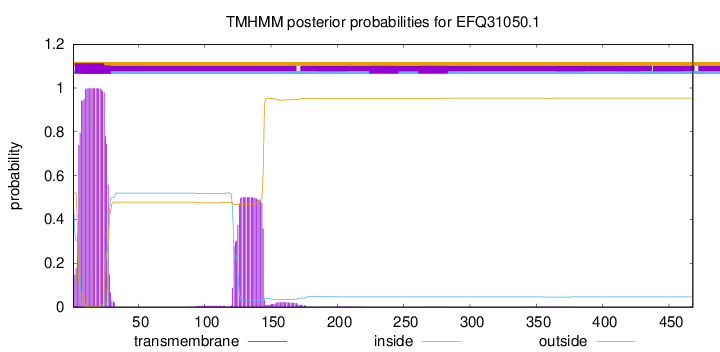
TMHMM annotations
Basic Information help
| Species | Colletotrichum graminicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Glomerellaceae; Colletotrichum; Colletotrichum graminicola | |||||||||||
| CAZyme ID | EFQ31050.1 | |||||||||||
| CAZy Family | GH146 | |||||||||||
| CAZyme Description | Chitobiosyldiphosphodolichol beta-mannosyltransferase [Source:UniProtKB/TrEMBL;Acc:E3QJL2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.142:10 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT33 | 45 | 461 | 8.6e-144 | 0.9905882352941177 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340843 | GT33_ALG1-like | 0.0 | 41 | 463 | 1 | 411 | chitobiosyldiphosphodolichol beta-mannosyltransferase and similar proteins. This family is most closely related to the GT33 family of glycosyltransferases. The yeast gene ALG1 has been shown to function as a mannosyltransferase that catalyzes the formation of dolichol pyrophosphate (Dol-PP)-GlcNAc2Man from GDP-Man and Dol-PP-Glc-NAc2, and participates in the formation of the lipid-linked precursor oligosaccharide for N-glycosylation. In humans ALG1 has been associated with the congenital disorders of glycosylation (CDG) designated as subtype CDG-Ik. |
| 215155 | PLN02275 | 7.28e-122 | 45 | 428 | 6 | 371 | transferase, transferring glycosyl groups |
| 340844 | GT4_UGDG-like | 1.56e-09 | 302 | 429 | 221 | 340 | UDP-Glc:1,2-diacylglycerol 3-a-glucosyltransferase and similar proteins. This family is most closely related to the GT1 family of glycosyltransferases. UDP-glucose-diacylglycerol glucosyltransferase (EC 2.4.1.337, UGDG; also known as 1,2-diacylglycerol 3-glucosyltransferase) catalyzes the transfer of glucose from UDP-glucose to 1,2-diacylglycerol forming 3-D-glucosyl-1,2-diacylglycerol. |
| 395425 | Glycos_transf_1 | 2.72e-08 | 302 | 435 | 22 | 149 | Glycosyl transferases group 1. Mutations in this domain of PIGA lead to disease (Paroxysmal Nocturnal haemoglobinuria). Members of this family transfer activated sugars to a variety of substrates, including glycogen, Fructose-6-phosphate and lipopolysaccharides. Members of this family transfer UDP, ADP, GDP or CMP linked sugars. The eukaryotic glycogen synthases may be distant members of this family. |
| 340847 | GT4_AmsD-like | 5.64e-08 | 282 | 445 | 191 | 335 | amylovoran biosynthesis glycosyltransferase AmsD and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. AmSD in Erwinia amylovora has been shown to be involved in the biosynthesis of amylovoran, the acidic exopolysaccharide acting as a virulence factor. This enzyme may be responsible for the formation of galactose alpha-1,6 linkages in amylovoran. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.19e-315 | 1 | 468 | 1 | 468 | |
| 2.16e-223 | 7 | 468 | 6 | 459 | |
| 5.84e-216 | 9 | 466 | 8 | 457 | |
| 1.67e-215 | 7 | 466 | 6 | 457 | |
| 2.37e-215 | 9 | 466 | 8 | 457 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.82e-111 | 27 | 466 | 19 | 446 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_01551 PE=3 SV=1 |
|
| 4.06e-93 | 46 | 463 | 27 | 420 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg1 PE=3 SV=2 |
|
| 2.22e-83 | 48 | 465 | 44 | 440 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=ALG1 PE=3 SV=1 |
|
| 5.61e-82 | 30 | 455 | 38 | 444 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=ALG1 PE=3 SV=1 |
|
| 6.53e-80 | 44 | 429 | 2 | 416 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Dictyostelium discoideum OX=44689 GN=alg1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
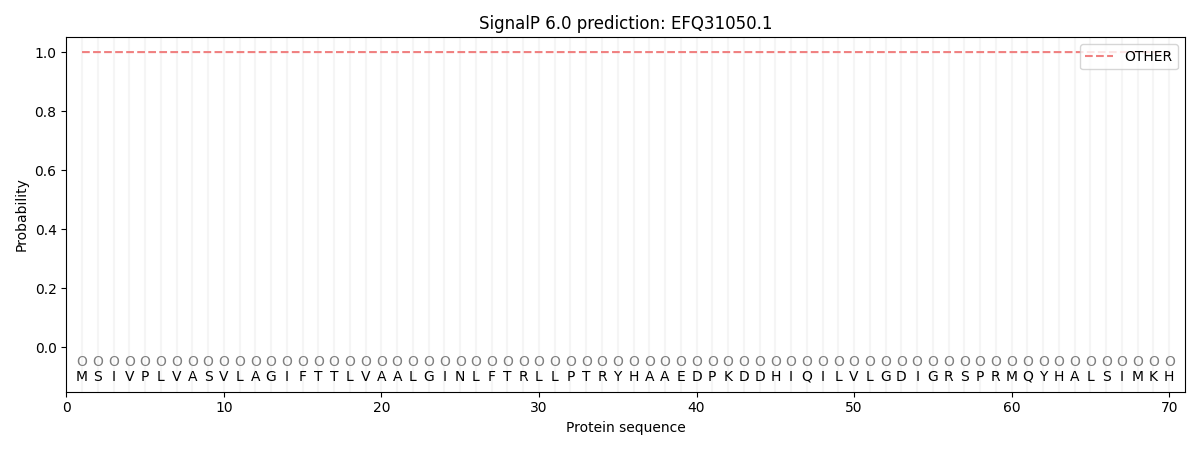
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000053 | 0.000000 |