You are browsing environment: FUNGIDB
CAZyme Information: EFQ30504.1
You are here: Home > Sequence: EFQ30504.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Colletotrichum graminicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Glomerellaceae; Colletotrichum; Colletotrichum graminicola | |||||||||||
| CAZyme ID | EFQ30504.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | Cell wall glucanase [Source:UniProtKB/TrEMBL;Acc:E3QI16] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 154 | 398 | 1.2e-23 | 0.9260450160771704 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.42e-73 | 100 | 404 | 4 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 1.07e-04 | 232 | 398 | 89 | 296 | Glycosyl hydrolases family 17. |
| 401378 | MSC | 0.005 | 313 | 385 | 97 | 163 | Man1-Src1p-C-terminal domain. MAN1 is an integral protein of the inner nuclear membrane which binds to chromatin associated proteins and plays a role in nuclear organisation. The C terminal nucleoplasmic region forms a DNA binding winged helix and binds to Smad. This C-terminal tail is also found in S. cerevisiae and is thought to consist of three conserved helices followed by two downstream strands. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.75e-174 | 5 | 401 | 3042 | 3448 | |
| 1.13e-102 | 143 | 398 | 80 | 333 | |
| 1.13e-102 | 143 | 398 | 80 | 333 | |
| 1.13e-102 | 143 | 398 | 80 | 333 | |
| 1.13e-102 | 143 | 398 | 80 | 333 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.62e-69 | 145 | 398 | 128 | 375 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
|
| 4.66e-66 | 145 | 398 | 135 | 382 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
|
| 6.02e-64 | 145 | 398 | 138 | 385 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 4.83e-43 | 145 | 397 | 297 | 546 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 1.10e-42 | 145 | 397 | 310 | 559 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
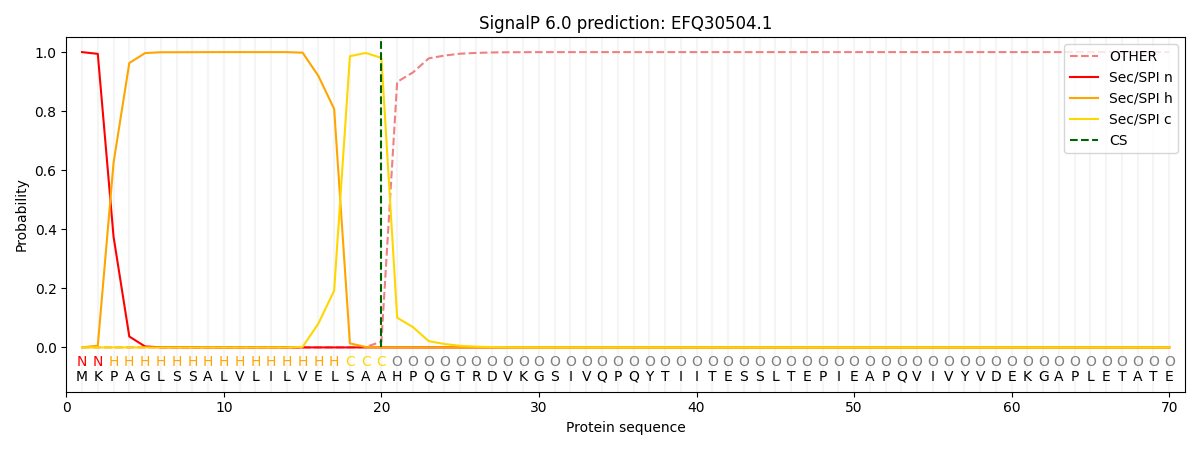
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000223 | 0.999749 | CS pos: 20-21. Pr: 0.9804 |
