You are browsing environment: FUNGIDB
CAZyme Information: EFQ29757.1
You are here: Home > Sequence: EFQ29757.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
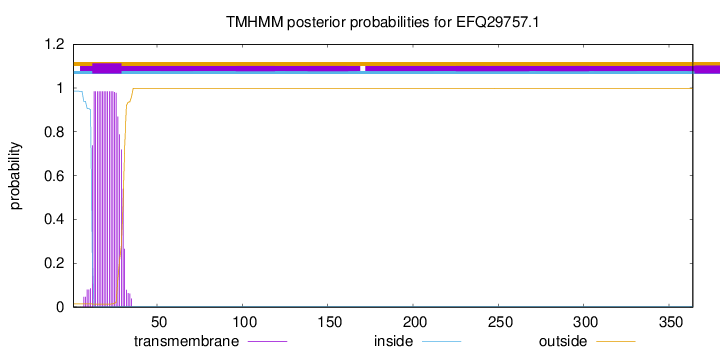
TMHMM annotations
Basic Information help
| Species | Colletotrichum graminicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Glomerellaceae; Colletotrichum; Colletotrichum graminicola | |||||||||||
| CAZyme ID | EFQ29757.1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Glycosyltransferase family 17 [Source:UniProtKB/TrEMBL;Acc:E3QFS2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT17 | 80 | 346 | 2.3e-72 | 0.9436619718309859 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398414 | Glyco_transf_17 | 1.26e-54 | 83 | 356 | 73 | 349 | Glycosyltransferase family 17. This family represents beta-1,4-mannosyl-glycoprotein beta-1,4-N-acetylglucosaminyltransferase (EC:2.4.1.144). This enzyme transfers the bisecting GlcNAc to the core mannose of complex N-glycans. The addition of this residue is regulated during development and has functional consequences for receptor signalling, cell adhesion, and tumor progression. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.64e-200 | 1 | 364 | 239 | 605 | |
| 2.21e-106 | 48 | 364 | 44 | 358 | |
| 1.43e-86 | 65 | 364 | 52 | 357 | |
| 5.74e-85 | 28 | 357 | 43 | 377 | |
| 1.71e-84 | 61 | 356 | 46 | 354 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.30e-09 | 48 | 346 | 175 | 499 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Homo sapiens OX=9606 GN=MGAT3 PE=1 SV=3 |
|
| 2.20e-08 | 48 | 346 | 179 | 503 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Rattus norvegicus OX=10116 GN=Mgat3 PE=1 SV=2 |
|
| 5.17e-08 | 48 | 346 | 179 | 503 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Mus musculus OX=10090 GN=Mgat3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999988 | 0.000018 |