You are browsing environment: FUNGIDB
CAZyme Information: EFQ27362.1
You are here: Home > Sequence: EFQ27362.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Colletotrichum graminicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Glomerellaceae; Colletotrichum; Colletotrichum graminicola | |||||||||||
| CAZyme ID | EFQ27362.1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase [Source:UniProtKB/TrEMBL;Acc:E3Q9I3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 38 | 214 | 1e-74 | 0.9943502824858758 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 3.87e-108 | 38 | 212 | 1 | 175 | Glycosyl hydrolases family 11. |
| 395595 | CBM_1 | 5.01e-12 | 251 | 279 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 5.73e-12 | 250 | 282 | 1 | 33 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.65e-135 | 1 | 218 | 11 | 228 | |
| 8.49e-134 | 1 | 282 | 1 | 281 | |
| 7.85e-131 | 1 | 282 | 1 | 286 | |
| 7.85e-131 | 1 | 282 | 1 | 286 | |
| 7.85e-131 | 1 | 282 | 1 | 286 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.60e-104 | 30 | 215 | 6 | 192 | High resolution structure of GH11 xylanase from Nectria haematococca [Fusarium vanettenii 77-13-4] |
|
| 1.04e-100 | 22 | 215 | 1 | 192 | Crystal Structure of a Xylanase at 1.56 Angstroem resolution [Fusarium oxysporum],5JRN_A Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution [Fusarium oxysporum f. sp. vasinfectum 25433] |
|
| 2.68e-95 | 28 | 215 | 1 | 189 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei] |
|
| 6.63e-95 | 30 | 218 | 4 | 193 | Chain A, endoxylanase 11A [Thermochaetoides thermophila],1XNK_B Chain B, endoxylanase 11A [Thermochaetoides thermophila] |
|
| 7.41e-95 | 34 | 215 | 6 | 188 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],4XQD_B Chain B, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],5ZF3_A Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose [Trichoderma reesei RUT C-30],5ZH0_A Crystal Structures of Endo-beta-1,4-xylanase II [Trichoderma reesei RUT C-30],5ZO0_A Neutron structure of xylanase at pD5.4 [Trichoderma reesei RUT C-30] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.08e-112 | 1 | 282 | 1 | 289 | Endo-1,4-beta-xylanase B OS=Phanerodontia chrysosporium OX=2822231 GN=xynB PE=1 SV=1 |
|
| 7.73e-111 | 1 | 215 | 1 | 218 | Endo-1,4-beta-xylanase I OS=Cochliobolus carbonum OX=5017 GN=XYL1 PE=1 SV=1 |
|
| 5.31e-104 | 1 | 215 | 1 | 229 | Endo-1,4-beta-xylanase 4 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL4 PE=3 SV=1 |
|
| 5.31e-104 | 1 | 215 | 1 | 229 | Endo-1,4-beta-xylanase 4 OS=Magnaporthe grisea OX=148305 GN=XYL4 PE=3 SV=2 |
|
| 6.36e-103 | 1 | 215 | 1 | 231 | Probable endo-1,4-beta-xylanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xlnA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
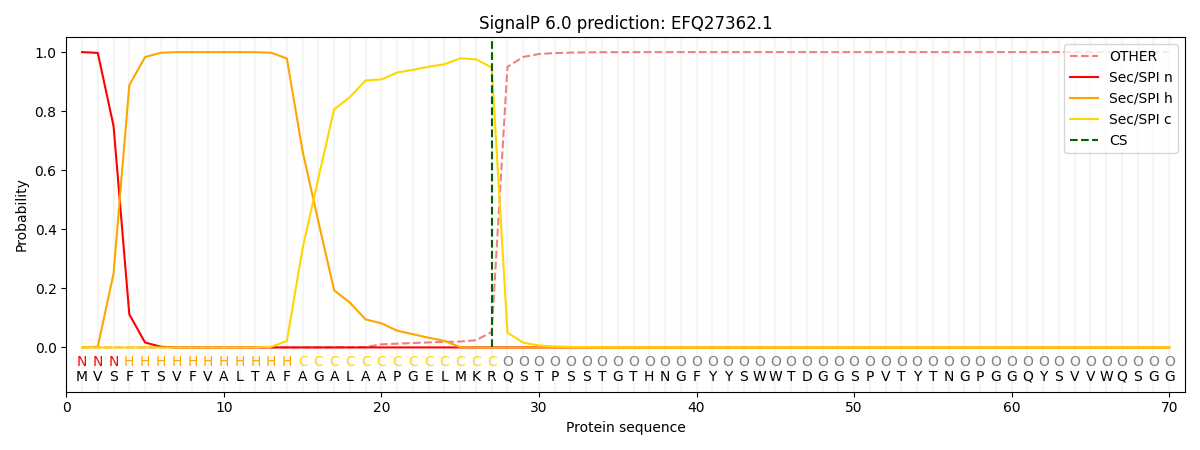
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000220 | 0.999738 | CS pos: 27-28. Pr: 0.9479 |
