You are browsing environment: FUNGIDB
CAZyme Information: EFQ25116.1
You are here: Home > Sequence: EFQ25116.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Colletotrichum graminicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Glomerellaceae; Colletotrichum; Colletotrichum graminicola | |||||||||||
| CAZyme ID | EFQ25116.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | Fungal cellulose binding domain-containing protein [Source:UniProtKB/TrEMBL;Acc:E3Q215] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA9 | 8 | 228 | 4.9e-58 | 0.9863636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410622 | LPMO_AA9 | 5.70e-69 | 20 | 242 | 1 | 216 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 9 (AA9). AA9 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs) involved in the cleavage of cellulose chains with oxidation of carbons C1 and/or C4 and C6. Activities include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54) and lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56). The family used to be called GH61 because weak endoglucanase activity had been demonstrated in some family members. |
| 397484 | Glyco_hydro_61 | 5.55e-60 | 21 | 233 | 1 | 211 | Glycosyl hydrolase family 61. Although weak endoglucanase activity has been demonstrated in several members of this family, they lack the clustered conserved catalytic acidic amino acids present in most glycoside hydrolases. Many members of this family lack measurable cellulase activity on their own, but enhance the activity of other cellulolytic enzymes. They are therefore unlikely to be true glycoside hydrolases. The subsrate-binding surface of this family is a flat Ig-like fold. |
| 237030 | kgd | 0.001 | 234 | 311 | 35 | 112 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 225689 | DedD | 0.002 | 234 | 311 | 66 | 144 | Cell division protein DedD (periplasmic protein involved in septation) [Cell cycle control, cell division, chromosome partitioning]. |
| 235906 | PRK07003 | 0.002 | 263 | 311 | 570 | 619 | DNA polymerase III subunit gamma/tau. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.41e-203 | 1 | 276 | 1 | 276 | |
| 1.96e-135 | 1 | 272 | 1 | 272 | |
| 5.02e-133 | 1 | 265 | 1 | 265 | |
| 1.58e-130 | 1 | 274 | 1 | 274 | |
| 1.58e-130 | 1 | 274 | 1 | 274 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.15e-26 | 20 | 244 | 1 | 221 | The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa [Neurospora crassa OR74A],4D7U_B The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa [Neurospora crassa OR74A],4D7V_A The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa [Neurospora crassa OR74A],4D7V_B The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa [Neurospora crassa OR74A] |
|
| 1.58e-23 | 20 | 244 | 1 | 217 | Chain A, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D [Phanerodontia chrysosporium],4B5Q_B Chain B, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D [Phanerodontia chrysosporium] |
|
| 1.48e-22 | 20 | 242 | 1 | 212 | Lytic polysaccharide monooxygenase 9F from Neurospora crassa, NcLPMO9F [Neurospora crassa],4QI8_B Lytic polysaccharide monooxygenase 9F from Neurospora crassa, NcLPMO9F [Neurospora crassa] |
|
| 1.80e-22 | 20 | 242 | 1 | 221 | Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKF_B Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKF_C Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKF_D Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKG_A Neurospora crassa polysaccharide monooxygenase 2 resting state [Neurospora crassa],5TKG_B Neurospora crassa polysaccharide monooxygenase 2 resting state [Neurospora crassa],5TKH_A Neurospora crassa polysaccharide monooxygenase 2 ascorbate treated [Neurospora crassa],5TKH_B Neurospora crassa polysaccharide monooxygenase 2 ascorbate treated [Neurospora crassa],5TKI_B Neurospora crassa polysaccharide monooxygenase 2 resting state joint X-ray/neutron refinement [Neurospora crassa] |
|
| 2.50e-22 | 20 | 242 | 1 | 221 | Neurospora crassa polysaccharide monooxygenase 2 resting state joint X-ray/neutron refinement [Neurospora crassa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.23e-31 | 1 | 250 | 1 | 240 | Probable endo-beta-1,4-glucanase D OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=eglD PE=3 SV=1 |
|
| 1.23e-26 | 4 | 244 | 5 | 237 | Probable endo-beta-1,4-glucanase D OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=eglD PE=3 SV=1 |
|
| 4.62e-26 | 4 | 244 | 5 | 237 | Probable endo-beta-1,4-glucanase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=eglD PE=3 SV=1 |
|
| 7.26e-26 | 1 | 244 | 1 | 234 | Probable endo-beta-1,4-glucanase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=eglD PE=3 SV=1 |
|
| 7.26e-26 | 1 | 244 | 1 | 234 | Probable endo-beta-1,4-glucanase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=eglD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
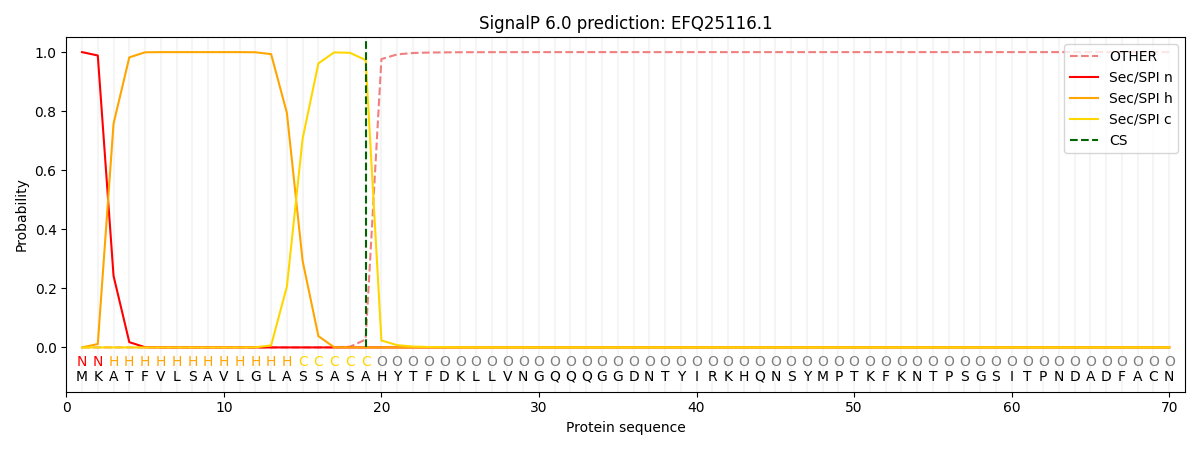
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000262 | 0.999684 | CS pos: 19-20. Pr: 0.9732 |
