You are browsing environment: FUNGIDB
CAZyme Information: EEU41078.1
You are here: Home > Sequence: EEU41078.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium vanettenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium vanettenii | |||||||||||
| CAZyme ID | EEU41078.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Amb_all domain-containing protein [Source:UniProtKB/TrEMBL;Acc:C7Z5D3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.2:31 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 82 | 262 | 9.8e-89 | 0.988950276243094 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 7.29e-66 | 82 | 266 | 2 | 190 | Amb_all domain. |
| 226384 | PelB | 7.82e-51 | 58 | 325 | 55 | 340 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 8.54e-41 | 91 | 262 | 29 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.16e-215 | 1 | 330 | 1 | 329 | |
| 1.36e-160 | 1 | 330 | 1 | 328 | |
| 1.36e-160 | 1 | 330 | 1 | 328 | |
| 1.36e-160 | 1 | 330 | 1 | 328 | |
| 1.36e-160 | 1 | 330 | 1 | 328 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.70e-42 | 43 | 264 | 6 | 248 | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
|
| 1.11e-32 | 37 | 240 | 4 | 247 | Chain A, PECTATE LYASE E [Dickeya chrysanthemi] |
|
| 6.11e-30 | 50 | 251 | 20 | 225 | Catalytic function and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
|
| 3.03e-27 | 42 | 239 | 15 | 257 | Chain A, Pectate lyase A [Dickeya chrysanthemi],1OOC_B Chain B, Pectate lyase A [Dickeya chrysanthemi],1PE9_A Chain A, Pectate lyase A [Dickeya chrysanthemi],1PE9_B Chain B, Pectate lyase A [Dickeya chrysanthemi] |
|
| 7.97e-27 | 42 | 239 | 15 | 257 | Chain A, Pectate lyase [Dickeya chrysanthemi],1JRG_B Chain B, Pectate lyase [Dickeya chrysanthemi],1JTA_A Chain A, pectate lyase A [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.79e-120 | 17 | 328 | 22 | 329 | Pectate lyase B OS=Colletotrichum gloeosporioides OX=474922 GN=PLB PE=3 SV=1 |
|
| 6.31e-108 | 1 | 328 | 1 | 326 | Probable pectate lyase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyB PE=3 SV=1 |
|
| 6.31e-108 | 1 | 328 | 1 | 326 | Probable pectate lyase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyB PE=3 SV=1 |
|
| 5.87e-106 | 1 | 328 | 1 | 326 | Pectate lyase plyB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyB PE=1 SV=1 |
|
| 1.06e-103 | 12 | 328 | 8 | 325 | Probable pectate lyase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
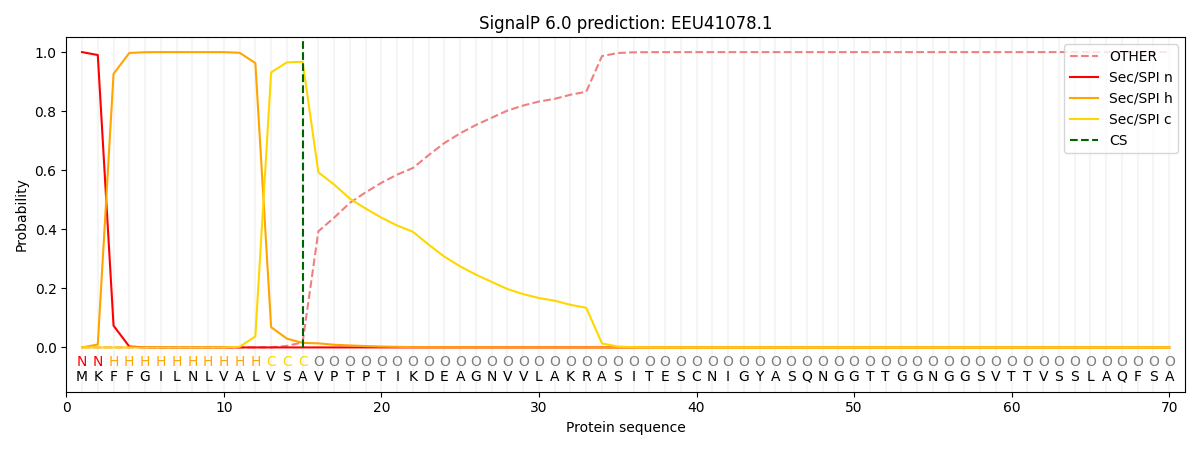
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999725 | CS pos: 15-16. Pr: 0.9673 |
