You are browsing environment: FUNGIDB
CAZyme Information: EEU40349.1
You are here: Home > Sequence: EEU40349.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
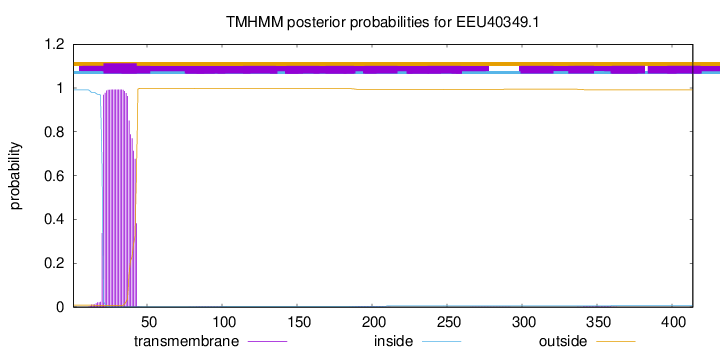
TMHMM annotations
Basic Information help
| Species | Fusarium vanettenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium vanettenii | |||||||||||
| CAZyme ID | EEU40349.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Cryptococcal mannosyltransferase 1-domain-containing protein [Source:UniProtKB/TrEMBL;Acc:C7Z4H7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT69 | 61 | 317 | 2.5e-84 | 0.9916317991631799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403053 | CAP59_mtransfer | 2.20e-117 | 61 | 312 | 1 | 225 | Cryptococcal mannosyltransferase 1. The capsule of pathogenic fungi is a complex polysaccharide whose formation is determined by a number of enzymes including, most importantly, alpha-1,3-mannosyltransferase 1, EC:2.4.1.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.23e-236 | 36 | 414 | 10 | 386 | |
| 1.81e-171 | 22 | 414 | 18 | 410 | |
| 2.22e-163 | 30 | 414 | 28 | 443 | |
| 1.65e-157 | 8 | 414 | 7 | 429 | |
| 3.24e-154 | 22 | 414 | 21 | 417 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
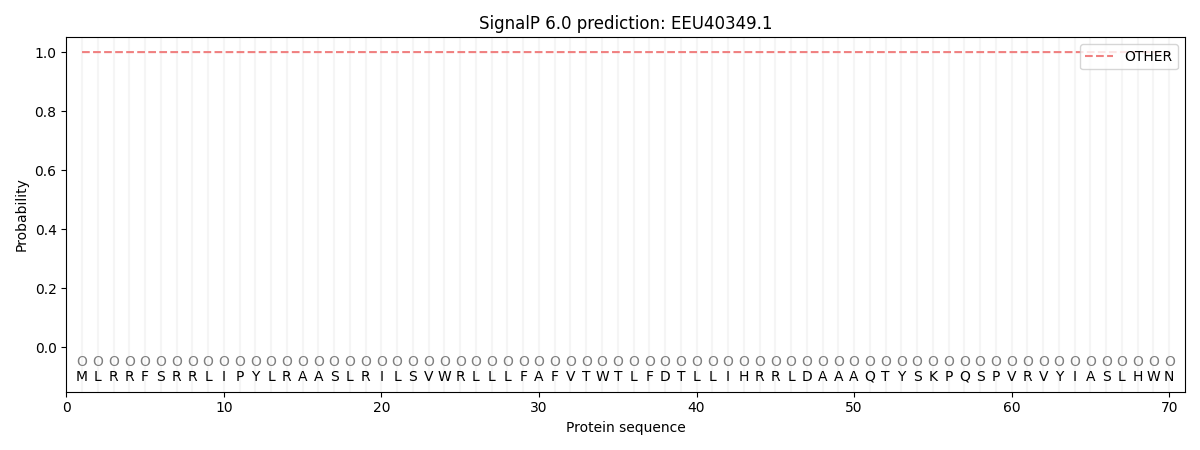
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000028 | 0.000005 |