You are browsing environment: FUNGIDB
CAZyme Information: EEU38641.1
You are here: Home > Sequence: EEU38641.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium vanettenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium vanettenii | |||||||||||
| CAZyme ID | EEU38641.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | Pectate lyase [Source:UniProtKB/TrEMBL;Acc:C7ZB71] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 31 | 216 | 2.4e-74 | 0.9896907216494846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 1.99e-96 | 25 | 219 | 2 | 198 | Pectate lyase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.48e-135 | 1 | 224 | 1 | 208 | |
| 2.15e-118 | 8 | 224 | 9 | 230 | |
| 2.31e-92 | 14 | 224 | 15 | 231 | |
| 1.61e-89 | 24 | 224 | 23 | 228 | |
| 1.18e-87 | 2 | 224 | 3 | 231 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.88e-27 | 38 | 162 | 7 | 132 | Crystal Structure Of Pectate Lyase From Bacillus Sp. Strain Ksm-P15. [Bacillus sp. KSM-P15] |
|
| 2.23e-19 | 37 | 162 | 11 | 135 | The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725],4EW9_B The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725] |
|
| 2.28e-19 | 37 | 162 | 12 | 136 | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii],3T9G_B The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii] |
|
| 1.99e-18 | 37 | 162 | 20 | 144 | C. bescii Family 3 pectate lyase double mutant K108A in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725],4Z03_B C. bescii Family 3 pectate lyase double mutant K108A in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725] |
|
| 1.99e-18 | 37 | 162 | 20 | 144 | C. bescii Family 3 pectate lyase double mutant K108A/E39Q in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725],4YZ0_B C. bescii Family 3 pectate lyase double mutant K108A/E39Q in complex with trigalacturonic acid [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.54e-86 | 15 | 224 | 12 | 232 | Probable pectate lyase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyF PE=3 SV=2 |
|
| 1.22e-85 | 16 | 216 | 13 | 218 | Probable pectate lyase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyF PE=3 SV=2 |
|
| 7.23e-85 | 4 | 224 | 1 | 232 | Probable pectate lyase F OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyF PE=3 SV=1 |
|
| 4.15e-84 | 7 | 224 | 4 | 224 | Probable pectate lyase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyF PE=3 SV=1 |
|
| 5.89e-84 | 16 | 224 | 13 | 224 | Probable pectate lyase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
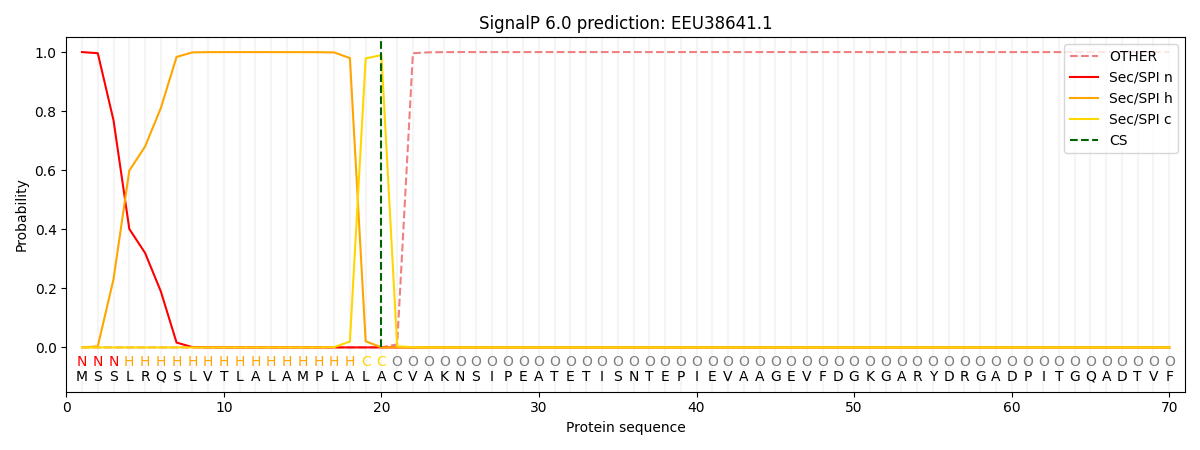
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000000 | 1.000059 | CS pos: 20-21. Pr: 0.9899 |
