You are browsing environment: FUNGIDB
CAZyme Information: EEU35841.1
You are here: Home > Sequence: EEU35841.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium vanettenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium vanettenii | |||||||||||
| CAZyme ID | EEU35841.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Sialidase domain-containing protein [Source:UniProtKB/TrEMBL;Acc:C7ZJF3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 42 | 346 | 1.5e-88 | 0.9674267100977199 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 271234 | Sialidase_non-viral | 4.38e-04 | 75 | 214 | 117 | 243 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 271234 | Sialidase_non-viral | 0.007 | 35 | 159 | 138 | 249 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.43e-265 | 1 | 376 | 1 | 357 | |
| 8.19e-147 | 8 | 376 | 15 | 391 | |
| 2.51e-144 | 23 | 376 | 34 | 396 | |
| 6.72e-126 | 21 | 376 | 30 | 392 | |
| 5.28e-120 | 23 | 376 | 30 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.74e-97 | 25 | 374 | 11 | 361 | Chain A, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],5M1Z_A Chain A, Exo-1,5-alpha-L-arabinofuranobiosidase [Fusarium graminearum] |
|
| 1.24e-96 | 25 | 374 | 11 | 361 | Chain A, Alpha-l-arabinofuranosidase [Fusarium graminearum] |
|
| 7.01e-96 | 25 | 374 | 11 | 361 | Chain A, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_B Chain B, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_C Chain C, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum] |
|
| 2.25e-94 | 25 | 374 | 11 | 361 | Chain A, ALPHA-L-ARABINOFURANOSIDASE [Fusarium graminearum] |
|
| 4.95e-93 | 26 | 374 | 7 | 351 | High resolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
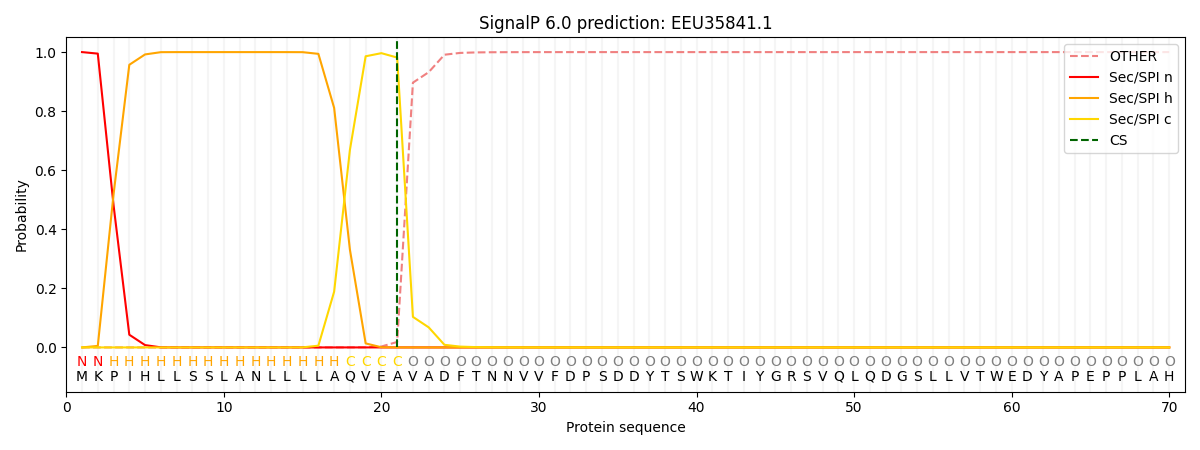
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000214 | 0.999787 | CS pos: 21-22. Pr: 0.9815 |
