You are browsing environment: FUNGIDB
CAZyme Information: EEQ35188.1
You are here: Home > Sequence: EEQ35188.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Microsporum canis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Arthrodermataceae; Microsporum; Microsporum canis | |||||||||||
| CAZyme ID | EEQ35188.1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | glyoxal oxidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA5 | 84 | 325 | 5.7e-67 | 0.42628774422735344 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 401164 | DUF1929 | 8.35e-22 | 222 | 312 | 6 | 91 | Domain of unknown function (DUF1929). Members of this family adopt a secondary structure consisting of a bundle of seven, mostly antiparallel, beta-strands surrounding a hydrophobic core. The 7 strands are arranged in 2 sheets, in a Greek-key topology. Their precise function, has not, as yet, been defined, though they are mostly found in sugar-utilising enzymes, such as galactose oxidase. |
| 199882 | E_set_GO_C | 2.39e-20 | 209 | 312 | 5 | 103 | C-terminal Early set domain associated with the catalytic domain of galactose oxidase. E or "early" set domains are associated with the catalytic domain of galactose oxidase at the C-terminal end. Galactose oxidase is an extracellular monomeric enzyme which catalyzes the stereospecific oxidation of a broad range of primary alcohol substrates and possesses a unique mononuclear copper site essential for catalyzing a two-electron transfer reaction during the oxidation of primary alcohols to corresponding aldehydes. The second redox active center necessary for the reaction was found to be situated at a tyrosine residue. The C-terminal domain of galactose oxidase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.17e-90 | 104 | 312 | 419 | 627 | |
| 2.31e-90 | 104 | 312 | 419 | 627 | |
| 1.58e-76 | 103 | 312 | 358 | 567 | |
| 8.41e-76 | 103 | 312 | 622 | 826 | |
| 1.34e-74 | 103 | 312 | 377 | 583 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.07e-08 | 113 | 312 | 456 | 637 | Chain A, Galactose oxidase [Fusarium graminearum] |
|
| 6.07e-08 | 113 | 312 | 456 | 637 | Chain A, Galactose oxidase [Fusarium graminearum] |
|
| 6.07e-08 | 113 | 312 | 456 | 637 | Chain A, Galactose oxidase [Fusarium graminearum] |
|
| 6.07e-08 | 113 | 312 | 456 | 637 | Glactose oxidase C383S mutant identified by directed evolution [Fusarium sp.] |
|
| 6.07e-08 | 113 | 312 | 456 | 637 | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.98e-28 | 104 | 312 | 409 | 614 | Aldehyde oxidase GLOX1 OS=Arabidopsis thaliana OX=3702 GN=GLOX1 PE=2 SV=1 |
|
| 3.78e-24 | 104 | 312 | 320 | 521 | Aldehyde oxidase GLOX OS=Vitis pseudoreticulata OX=231512 GN=GLOX PE=2 SV=1 |
|
| 8.37e-21 | 103 | 325 | 716 | 912 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 2.88e-16 | 104 | 312 | 392 | 593 | Putative aldehyde oxidase Art an 7 OS=Artemisia annua OX=35608 PE=1 SV=1 |
|
| 1.13e-12 | 120 | 314 | 365 | 551 | Aldehyde oxidase GLOX OS=Phanerodontia chrysosporium OX=2822231 GN=GLX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
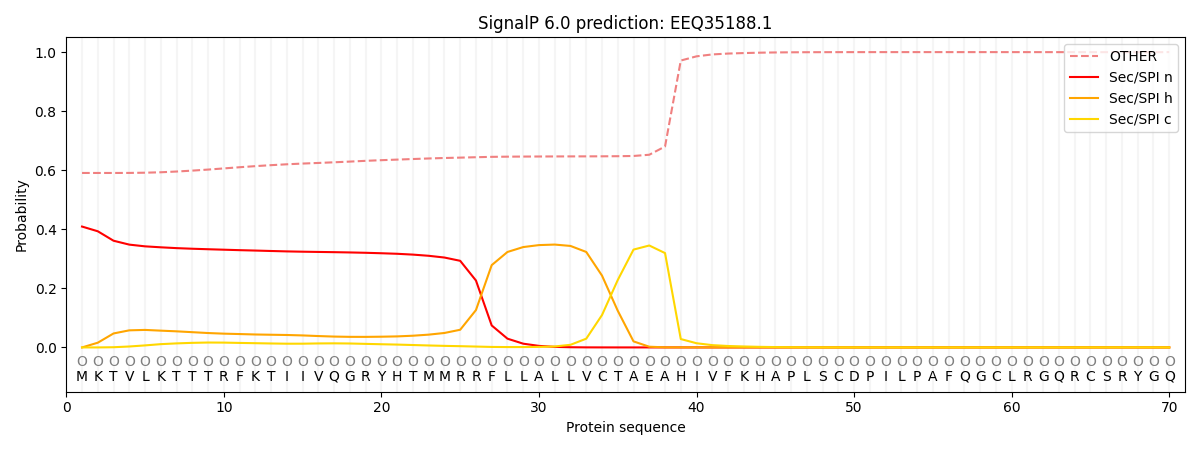
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.594980 | 0.405010 |
