You are browsing environment: FUNGIDB
CAZyme Information: EEP75320.1
You are here: Home > Sequence: EEP75320.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Uncinocarpus reesii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Onygenaceae; Uncinocarpus; Uncinocarpus reesii | |||||||||||
| CAZyme ID | EEP75320.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | MUTSd domain-containing protein [Source:UniProtKB/TrEMBL;Acc:C4JKV1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 18 | 207 | 6.9e-67 | 0.9685863874345549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223327 | MutS | 3.27e-21 | 449 | 596 | 176 | 317 | DNA mismatch repair ATPase MutS [Replication, recombination and repair]. |
| 398732 | MutS_III | 4.70e-15 | 544 | 596 | 1 | 52 | MutS domain III. This domain is found in proteins of the MutS family (DNA mismatch repair proteins) and is found associated with pfam00488, pfam05188, pfam01624 and pfam05190. The MutS family of proteins is named after the Salmonella typhimurium MutS protein involved in mismatch repair; other members of the family included the eukaryotic MSH 1,2,3, 4,5 and 6 proteins. These have various roles in DNA repair and recombination. Human MSH has been implicated in non-polyposis colorectal carcinoma (HNPCC) and is a mismatch binding protein. The aligned region corresponds with domain III, which is central to the structure of Thermus aquaticus MutS as characterized in. |
| 214710 | MUTSd | 3.80e-11 | 561 | 596 | 1 | 36 | DNA-binding domain of DNA mismatch repair MUTS family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.16e-78 | 11 | 222 | 11 | 222 | |
| 6.36e-75 | 11 | 220 | 11 | 219 | |
| 6.36e-75 | 11 | 220 | 11 | 219 | |
| 6.36e-75 | 11 | 220 | 11 | 219 | |
| 6.36e-75 | 11 | 220 | 11 | 219 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.13e-38 | 18 | 206 | 1 | 201 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
|
| 6.95e-33 | 452 | 596 | 209 | 358 | human MutSalpha (MSH2/MSH6) bound to ADP and a G T mispair [Homo sapiens],2O8C_A human MutSalpha (MSH2/MSH6) bound to ADP and an O6-methyl-guanine T mispair [Homo sapiens],2O8D_A human MutSalpha (MSH2/MSH6) bound to ADP and a G dU mispair [Homo sapiens],2O8F_A human MutSalpha (MSH2/MSH6) bound to DNA with a single base T insert [Homo sapiens],3THW_A Human MutSbeta complexed with an IDL of 4 bases (Loop4) and ADP [Homo sapiens],3THX_A Human MutSbeta complexed with an IDL of 3 bases (Loop3) and ADP [Homo sapiens],3THY_A Human MutSbeta complexed with an IDL of 2 bases (Loop2) and ADP [Homo sapiens],3THZ_A Human MutSbeta complexed with an IDL of 6 bases (Loop6) and ADP [Homo sapiens] |
|
| 6.95e-33 | 452 | 596 | 209 | 358 | Chain A, DNA mismatch repair protein Msh2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.21e-66 | 449 | 596 | 200 | 347 | DNA mismatch repair protein msh-2 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=msh-2 PE=3 SV=1 |
|
| 3.57e-32 | 452 | 596 | 209 | 358 | DNA mismatch repair protein Msh2 OS=Chlorocebus aethiops OX=9534 GN=MSH2 PE=2 SV=1 |
|
| 3.58e-32 | 452 | 596 | 209 | 358 | DNA mismatch repair protein Msh2 OS=Homo sapiens OX=9606 GN=MSH2 PE=1 SV=1 |
|
| 3.58e-32 | 452 | 596 | 209 | 358 | DNA mismatch repair protein Msh2 OS=Bos taurus OX=9913 GN=MSH2 PE=2 SV=1 |
|
| 2.63e-31 | 452 | 596 | 209 | 358 | DNA mismatch repair protein Msh2 OS=Mus musculus OX=10090 GN=Msh2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
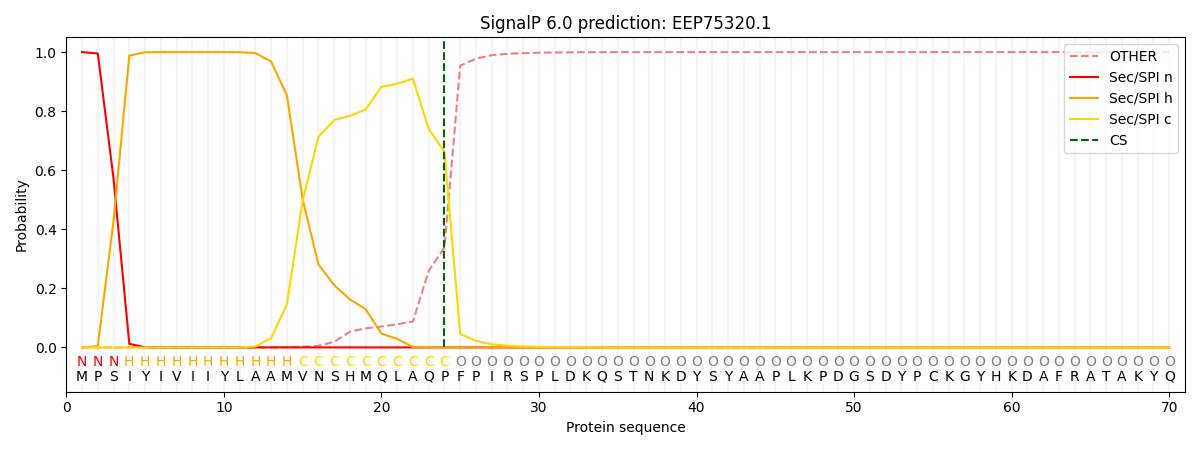
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000229 | 0.999748 | CS pos: 24-25. Pr: 0.6619 |
