You are browsing environment: FUNGIDB
CAZyme Information: EDK40210.2
You are here: Home > Sequence: EDK40210.2
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
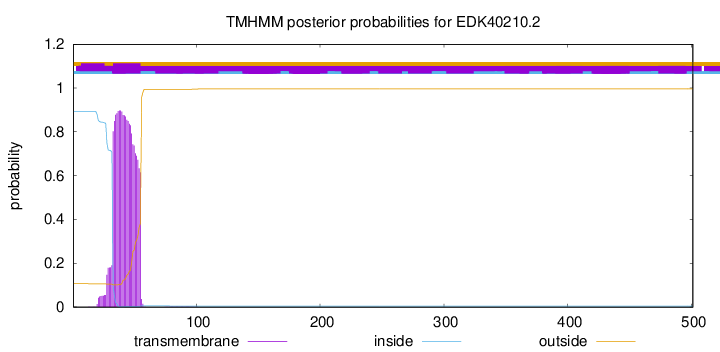
TMHMM annotations
Basic Information help
| Species | Meyerozyma guilliermondii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Meyerozyma; Meyerozyma guilliermondii | |||||||||||
| CAZyme ID | EDK40210.2 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA2 | 225 | 477 | 1.7e-58 | 0.9764705882352941 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173825 | ascorbate_peroxidase | 4.71e-121 | 215 | 481 | 6 | 253 | Ascorbate peroxidases and cytochrome C peroxidases. Ascorbate peroxidases are a subgroup of heme-dependent peroxidases of the plant superfamily that share a heme prosthetic group and catalyze a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. Along with related catalase-peroxidases, ascorbate peroxidases belong to class I of the plant superfamily. Ascorbate peroxidases are found in the chloroplasts and/or cytosol of algae and plants, where they have been shown to control the concentration of lethal hydrogen peroxide molecules. The yeast cytochrome c peroxidase is a divergent member of the family; it forms a complex with cytochrome c to catalyze the reduction of hydrogen peroxide to water. |
| 178218 | PLN02608 | 3.60e-65 | 218 | 479 | 7 | 245 | L-ascorbate peroxidase |
| 178467 | PLN02879 | 1.16e-57 | 244 | 477 | 34 | 246 | L-ascorbate peroxidase |
| 395089 | peroxidase | 4.04e-55 | 225 | 458 | 1 | 187 | Peroxidase. |
| 223453 | KatG | 8.57e-55 | 239 | 458 | 90 | 397 | Catalase (peroxidase I) [Inorganic ion transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.43e-94 | 223 | 485 | 7 | 268 | |
| 1.73e-93 | 222 | 486 | 19 | 287 | |
| 8.64e-91 | 225 | 486 | 22 | 287 | |
| 8.64e-91 | 225 | 486 | 22 | 287 | |
| 8.64e-91 | 225 | 486 | 22 | 287 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.47e-72 | 243 | 483 | 25 | 263 | Structure of Leishmania major peroxidase D211N mutant [Leishmania major],5AMM_B Structure of Leishmania major peroxidase D211N mutant [Leishmania major] |
|
| 4.59e-72 | 243 | 483 | 25 | 263 | Crystal Structure of the Leishmania Major Peroxidase-Cytochrome C Complex [Leishmania major] |
|
| 4.89e-72 | 243 | 483 | 25 | 263 | Structure of Leishmania major peroxidase D211R mutant (high res) [Leishmania major],5ALA_A Structure of Leishmania major peroxidase D211R mutant (low res) [Leishmania major],5ALA_B Structure of Leishmania major peroxidase D211R mutant (low res) [Leishmania major] |
|
| 5.04e-72 | 243 | 483 | 26 | 264 | The Crystal Structure of Leishmania major Peroxidase [Leishmania major strain Friedlin],3RIV_B The Crystal Structure of Leishmania major Peroxidase [Leishmania major strain Friedlin] |
|
| 1.55e-70 | 243 | 483 | 26 | 264 | The Crystal Structure of Leishmania major Peroxidase mutant C197T [Leishmania major strain Friedlin],3RIW_B The Crystal Structure of Leishmania major Peroxidase mutant C197T [Leishmania major strain Friedlin] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-137 | 171 | 490 | 92 | 425 | Putative heme-binding peroxidase OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=DEHA2G12166g PE=3 SV=3 |
|
| 4.86e-124 | 224 | 481 | 32 | 289 | Putative heme-binding peroxidase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CCP2 PE=3 SV=2 |
|
| 9.65e-95 | 223 | 485 | 7 | 268 | Putative cytochrome c peroxidase, mitochondrial OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=YALI0D04268g PE=3 SV=1 |
|
| 6.82e-90 | 225 | 487 | 22 | 288 | Putative heme-binding peroxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FGRRES_10606 PE=3 SV=1 |
|
| 1.84e-87 | 223 | 486 | 8 | 277 | Putative heme-binding peroxidase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=AN5440 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999997 | 0.000003 |