You are browsing environment: FUNGIDB
CAZyme Information: EDK37078.2
You are here: Home > Sequence: EDK37078.2
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Meyerozyma guilliermondii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Meyerozyma; Meyerozyma guilliermondii | |||||||||||
| CAZyme ID | EDK37078.2 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 265 | 510 | 1.4e-22 | 0.9292604501607717 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.36e-74 | 211 | 510 | 4 | 301 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 6.07e-07 | 286 | 510 | 38 | 297 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.27e-127 | 237 | 514 | 262 | 539 | |
| 1.28e-126 | 222 | 515 | 139 | 431 | |
| 3.59e-123 | 237 | 514 | 273 | 550 | |
| 9.71e-120 | 248 | 514 | 36 | 302 | |
| 2.36e-113 | 226 | 515 | 126 | 411 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.40e-09 | 257 | 511 | 39 | 283 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.96e-79 | 252 | 509 | 133 | 385 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 5.09e-77 | 255 | 510 | 126 | 376 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
|
| 1.79e-76 | 252 | 509 | 130 | 382 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
|
| 2.60e-36 | 257 | 508 | 304 | 553 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 1.92e-35 | 254 | 509 | 280 | 537 | Probable family 17 glucosidase SCW11 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW11 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
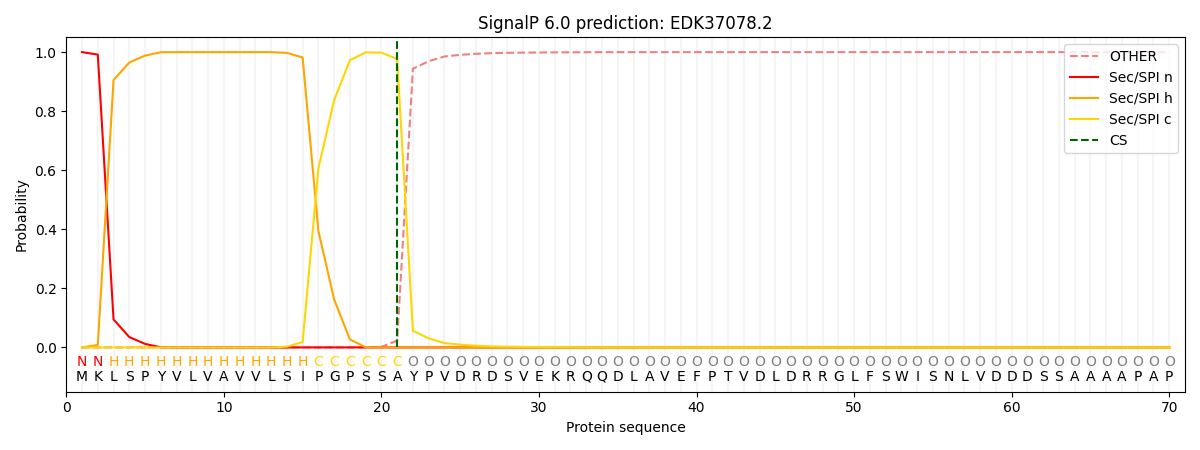
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000264 | 0.999701 | CS pos: 21-22. Pr: 0.9769 |
