You are browsing environment: FUNGIDB
CAZyme Information: EAW25175.1
You are here: Home > Sequence: EAW25175.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
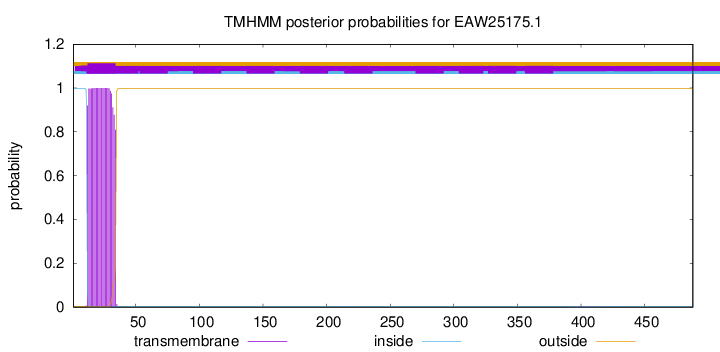
TMHMM annotations
Basic Information help
| Species | Aspergillus fischeri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fischeri | |||||||||||
| CAZyme ID | EAW25175.1 | |||||||||||
| CAZy Family | GT69 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT61 | 295 | 414 | 5.6e-26 | 0.957983193277311 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398330 | DUF563 | 1.04e-27 | 210 | 438 | 4 | 203 | Protein of unknown function (DUF563). Family of uncharacterized proteins. |
| 226845 | COG4421 | 2.08e-07 | 308 | 454 | 189 | 341 | Capsular polysaccharide biosynthesis protein [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.89e-179 | 6 | 487 | 7 | 508 | |
| 2.08e-152 | 6 | 487 | 6 | 491 | |
| 9.05e-135 | 60 | 488 | 60 | 501 | |
| 5.13e-134 | 60 | 488 | 60 | 501 | |
| 1.36e-132 | 61 | 488 | 58 | 495 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.96e-11 | 257 | 449 | 158 | 361 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9J_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_A Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_D Chain D, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_E Chain E, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9L_A Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9L_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus] |
|
| 4.55e-09 | 257 | 434 | 148 | 334 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 6.06e-09 | 257 | 434 | 152 | 338 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 1.39e-08 | 257 | 434 | 148 | 334 | Chain C, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 1.85e-08 | 257 | 434 | 152 | 338 | Chain D, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.96e-22 | 43 | 416 | 49 | 431 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Drosophila melanogaster OX=7227 GN=Eogt PE=1 SV=1 |
|
| 2.34e-14 | 305 | 466 | 333 | 491 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Rattus norvegicus OX=10116 GN=Eogt PE=2 SV=1 |
|
| 3.11e-14 | 305 | 466 | 333 | 491 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Mus musculus OX=10090 GN=Eogt PE=1 SV=1 |
|
| 7.41e-14 | 305 | 466 | 341 | 499 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Gallus gallus OX=9031 GN=EOGT PE=2 SV=2 |
|
| 1.70e-13 | 193 | 466 | 230 | 491 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Bos taurus OX=9913 GN=EOGT PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
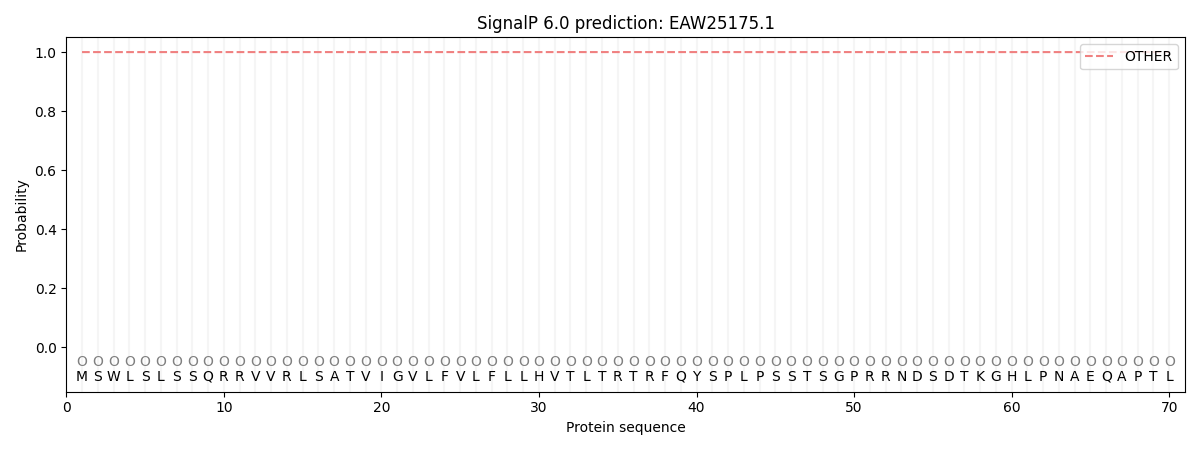
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000049 | 0.000001 |