You are browsing environment: FUNGIDB
CAZyme Information: EAW15612.1
Basic Information
help
| Species |
Aspergillus fischeri
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fischeri
|
| CAZyme ID |
EAW15612.1
|
| CAZy Family |
GT90 |
| CAZyme Description |
conserved hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 464 |
DS027698|CGC33 |
51420.45 |
7.1250 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AfischeriNRRL181 |
10668 |
331117 |
278 |
10390
|
|
| Gene Location |
Start: 715677; End:717223 Strand: + |
| MKTTIFASLL LAGTALGNPV HQKRDSVTAQ VNFANNTGKP QHLASGILYG LPDTPNQIPT | 60 |
| KFYTDMGFNY NRAGGAQVPA PGRGWIWGLN EYKTRFASAL SNYKTSRQHG AKFIFLIHDL | 120 |
| WGADGTQNSS APYPGDNGDW SSWDNYLTHL LSDIKANSMT DGLIIDIWNE PDLTYFWNRD | 180 |
| QTQYLQMWGR TFHRLRSELA GVELSGPASA GEPLPSNNWW KNWASFAATN KSIPDQYAWH | 240 |
| MEGGGGDLLS AQAGLVYWQK TYGLPGRPIN INEYAVLDEE VPAGSAWWIG QLERINAVGL | 300 |
| RGNWLSGWKL HDLLASLLSK PNADNSNYDP KGTGYFPNGD YQVYKYYNLN MTGHRVGTLP | 360 |
| SSDLKLDAYA TVGDDRVARV LVGVRIAKGT WQLQLNKLSA LGLPTSGTLN VHTWGFPVAS | 420 |
| NVHYGRVDGP KDLGWYGHAY SGDSVTFPVY QTDTTTAYAF EFKI | 464 |
No EC number prediction in EAW15612.1.
EAW15612.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5U22_A |
3.74e-14 |
142 |
363 |
119 |
324 |
Structure of N2152 from Neocallimastix frontalis [Neocallimastix frontalis] |
EAW15612.1 has no Swissprot hit.
This protein is predicted as SP
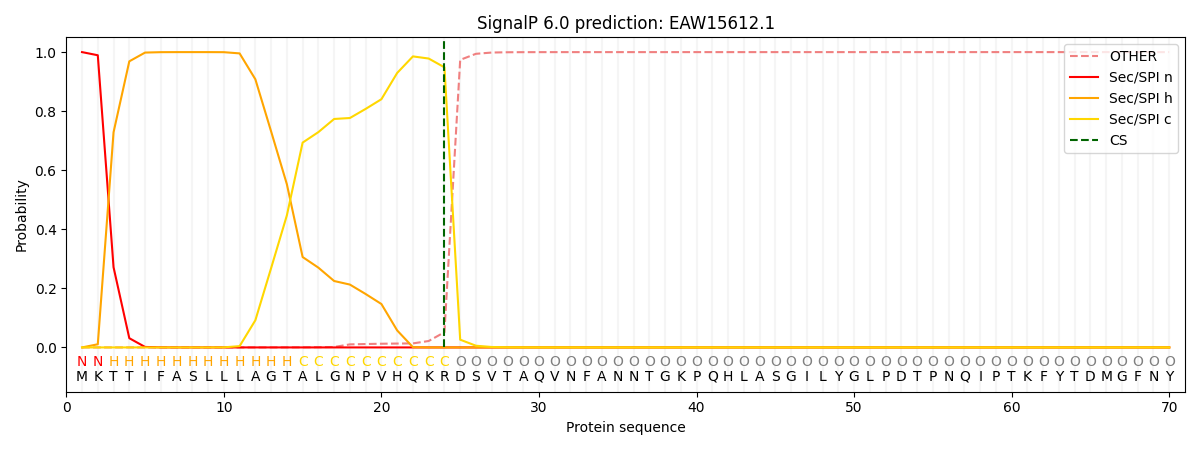
| Other |
SP_Sec_SPI |
CS Position |
| 0.000365 |
0.999606 |
CS pos: 24-25. Pr: 0.9485 |
There is no transmembrane helices in EAW15612.1.

