You are browsing environment: FUNGIDB
CAZyme Information: EAQ89046.1
You are here: Home > Sequence: EAQ89046.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
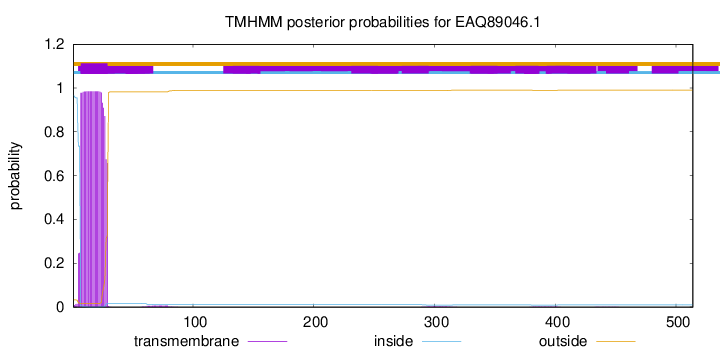
TMHMM annotations
Basic Information help
| Species | Chaetomium globosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Chaetomiaceae; Chaetomium; Chaetomium globosum | |||||||||||
| CAZyme ID | EAQ89046.1 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | Glyco_hydro_35 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:Q2H6Q0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.23:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 34 | 348 | 4e-89 | 0.990228013029316 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 4.90e-85 | 33 | 349 | 1 | 316 | Glycosyl hydrolases family 35. |
| 166698 | PLN03059 | 8.06e-34 | 26 | 345 | 29 | 336 | beta-galactosidase; Provisional |
| 224786 | GanA | 4.93e-23 | 30 | 183 | 4 | 162 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 1.40e-07 | 50 | 183 | 4 | 139 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| 404274 | BetaGal_dom4_5 | 6.78e-04 | 419 | 470 | 35 | 89 | Beta-galactosidase jelly roll domain. This domain is found in beta galactosidase enzymes. It has a jelly roll fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.92e-200 | 20 | 514 | 30 | 638 | |
| 2.15e-143 | 25 | 509 | 29 | 634 | |
| 3.75e-143 | 22 | 509 | 27 | 641 | |
| 2.08e-141 | 15 | 509 | 23 | 648 | |
| 1.17e-123 | 23 | 467 | 25 | 588 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.33e-78 | 15 | 497 | 16 | 604 | Galactanase BT0290 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 4.18e-73 | 25 | 484 | 6 | 572 | Crystal structure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.11e-65 | 28 | 485 | 19 | 578 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 3.85e-58 | 29 | 489 | 20 | 610 | Chain A, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
|
| 1.37e-54 | 29 | 489 | 13 | 602 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.31e-76 | 22 | 374 | 20 | 377 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
|
| 7.21e-67 | 34 | 501 | 56 | 623 | Beta-galactosidase-1-like protein 3 OS=Rattus norvegicus OX=10116 GN=Glb1l3 PE=2 SV=1 |
|
| 1.54e-66 | 33 | 484 | 53 | 613 | Beta-galactosidase-1-like protein 2 OS=Mus musculus OX=10090 GN=Glb1l2 PE=1 SV=1 |
|
| 1.43e-65 | 34 | 501 | 56 | 623 | Beta-galactosidase-1-like protein 3 OS=Mus musculus OX=10090 GN=Glb1l3 PE=1 SV=1 |
|
| 5.96e-64 | 33 | 484 | 53 | 613 | Beta-galactosidase-1-like protein 2 OS=Homo sapiens OX=9606 GN=GLB1L2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998232 | 0.001788 |