You are browsing environment: FUNGIDB
CAZyme Information: EAQ84293.1
You are here: Home > Sequence: EAQ84293.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Chaetomium globosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Chaetomiaceae; Chaetomium; Chaetomium globosum | |||||||||||
| CAZyme ID | EAQ84293.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | FAD-binding PCMH-type domain-containing protein [Source:UniProtKB/TrEMBL;Acc:Q2GMV7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 158 | 371 | 2.4e-42 | 0.42358078602620086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 9.09e-11 | 162 | 306 | 3 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 223354 | GlcD | 3.71e-10 | 167 | 591 | 39 | 450 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 369468 | MFS_1 | 1.13e-09 | 706 | 956 | 30 | 257 | Major Facilitator Superfamily. |
| 340883 | MFS_MdtG_SLC18_like | 2.12e-09 | 705 | 969 | 28 | 268 | bacterial MdtG-like and eukaryotic solute carrier 18 (SLC18) family of the Major Facilitator Superfamily of transporters. This family is composed of eukaryotic solute carrier 18 (SLC18) family transporters and related bacterial multidrug resistance (MDR) transporters including several proteins from Escherichia coli such as multidrug resistance protein MdtG, from Bacillus subtilis such as multidrug resistance proteins 1 (Bmr1) and 2 (Bmr2), and from Staphylococcus aureus such as quinolone resistance protein NorA. The family also includes Escherichia coli arabinose efflux transporters YfcJ and YhhS. MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. The SLC18 transporter family includes vesicular monoamine transporters (VAT1 and VAT2), vesicular acetylcholine transporter (VAChT), and SLC18B1, which is proposed to be a vesicular polyamine transporter (VPAT). The MdtG/SLC18 family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 369658 | BBE | 3.42e-06 | 554 | 593 | 1 | 40 | Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.64e-41 | 59 | 603 | 500 | 1055 | |
| 8.64e-41 | 59 | 603 | 500 | 1055 | |
| 3.07e-14 | 163 | 591 | 66 | 484 | |
| 6.40e-14 | 139 | 591 | 37 | 478 | |
| 7.46e-14 | 161 | 337 | 165 | 332 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.63e-81 | 47 | 608 | 16 | 569 | Crystal structure of VAO-type flavoprotein MtVAO615 at pH 7.5 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_A Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_B Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 4.66e-54 | 59 | 608 | 30 | 593 | Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_B Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_C Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_D Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 2.68e-16 | 161 | 341 | 54 | 228 | Crystal structure of Phl p 4, a grass pollen allergen with glucose dehydrogenase activity [Phleum pratense],3TSJ_A Crystal structure of Phl p 4, a grass pollen allergen with glucose dehydrogenase activity [Phleum pratense],3TSJ_B Crystal structure of Phl p 4, a grass pollen allergen with glucose dehydrogenase activity [Phleum pratense] |
|
| 2.68e-16 | 161 | 341 | 54 | 228 | Phl p 4 I153V variant, a glucose oxidase [Phleum pratense],4PWB_A Phl p 4 I153V variant, a glucose oxidase, pressurized with Xenon [Phleum pratense] |
|
| 3.55e-16 | 161 | 341 | 54 | 228 | Phl p 4 I153V N158H variant, a glucose oxidase [Phleum pratense],4PWC_A Phl p 4 I153V N158H variant, a glucose oxidase, 3.5 M NaBr soak [Phleum pratense] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.53e-109 | 46 | 608 | 14 | 558 | FAD-linked oxidoreductase ZEB1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ZEB1 PE=2 SV=2 |
|
| 1.46e-94 | 41 | 607 | 21 | 567 | FAD-linked oxidoreductase sor8 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=sor8 PE=3 SV=1 |
|
| 2.20e-88 | 54 | 608 | 23 | 562 | FAD-linked oxidoreductase patO OS=Penicillium expansum OX=27334 GN=patO PE=1 SV=1 |
|
| 2.75e-85 | 59 | 608 | 29 | 563 | FAD-linked oxidoreductase patO OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=patO PE=1 SV=1 |
|
| 1.21e-81 | 46 | 602 | 14 | 575 | FAD-linked oxidoreductase orf1 OS=Neocamarosporium betae OX=1979465 GN=orf1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
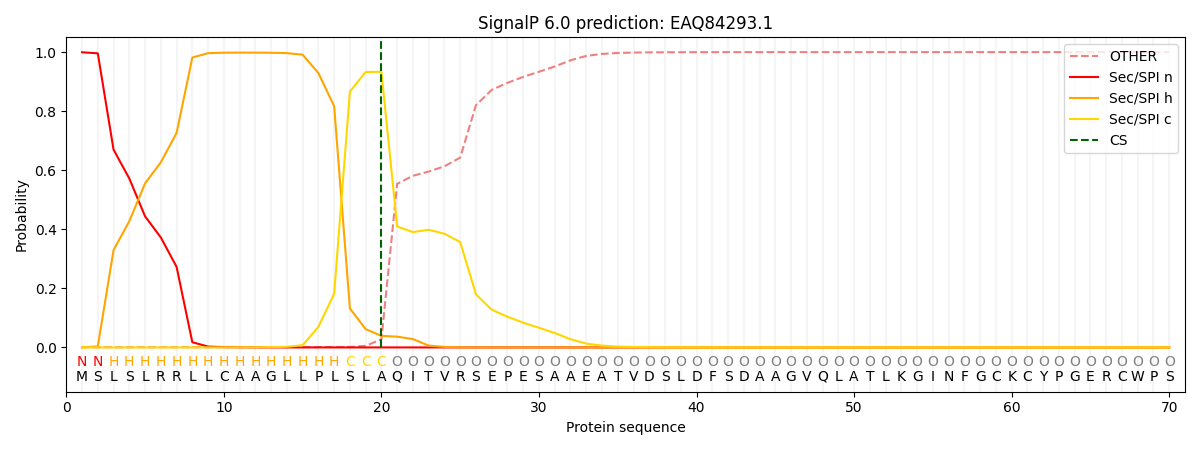
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001712 | 0.998261 | CS pos: 20-21. Pr: 0.9333 |
TMHMM Annotations download full data without filtering help
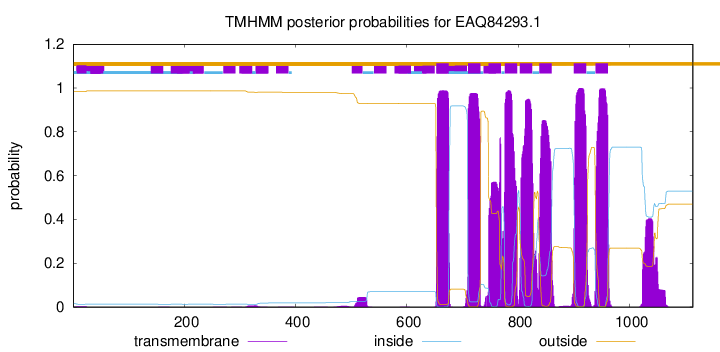
| Start | End |
|---|---|
| 653 | 675 |
| 710 | 732 |
| 747 | 769 |
| 776 | 798 |
| 803 | 825 |
| 838 | 860 |
| 900 | 922 |
| 939 | 961 |
