You are browsing environment: FUNGIDB
CAZyme Information: CXQ87_003106-t45_1-p1
You are here: Home > Sequence: CXQ87_003106-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] duobushaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] duobushaemulonis | |||||||||||
| CAZyme ID | CXQ87_003106-t45_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 7 | 372 | 9e-98 | 0.9385474860335196 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 1.37e-167 | 10 | 362 | 1 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 240537 | MATE_eukaryotic | 5.61e-149 | 525 | 957 | 1 | 431 | Eukaryotic members of the multidrug and toxic compound extrusion (MATE) family. The integral membrane proteins from the MATE family are involved in exporting metabolites across the cell membrane and are responsible for multidrug resistance (MDR) in many bacteria and animals. MATE has also been identified as a large multigene family in plants, where the proteins are linked to disease resistance. A number of family members are involved in the synthesis of peptidoglycan components in bacteria. This subfamily, which is restricted to eukaryotes, contains vertebrate solute transporters responsible for secretion of cationic drugs across the brush border membranes, yeast proteins located in the vacuole membrane, and plant proteins involved in disease resistance and iron homeostatis under osmotic stress. |
| 223608 | NorM | 3.81e-95 | 517 | 963 | 7 | 452 | Na+-driven multidrug efflux pump [Defense mechanisms]. |
| 240536 | MATE_NorM_like | 1.72e-74 | 525 | 945 | 1 | 423 | Subfamily of the multidrug and toxic compound extrusion (MATE)-like proteins similar to Vibrio cholerae NorM. The integral membrane proteins from the MATE family are involved in exporting metabolites across the cell membrane and are responsible for multidrug resistance (MDR) in many bacteria and animals. This subfamily includes Vibrio cholerae NorM and functions most likely as a multidrug efflux pump, removing xenobiotics from the interior of the cell. The pump utilizes a cation gradient across the membrane to facilitate the export process. NorM appears to bind monovalent cations in an outward-facing conformation and may subsequently cycle through an inward-facing and outward-facing conformation to capture and release its substrate. |
| 273273 | matE | 1.18e-60 | 535 | 872 | 1 | 338 | putative efflux protein, MATE family. The Multi Antimicrobial Extrusion (MATE) Family (TC 2.A.66) The MATE family consists of probable efflux proteins including a functionally characterized multi drug efflux system from Vibrio parahaemolyticus, a putative ethionine resistance protein of Saccharomyces cerevisiae, and the functionally uncharacterized DNA damage-inducible protein F (DinF) of E. coli. These proteins have 12 probable TMS. [Transport and binding proteins, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 975 | 1637 | 2644 | |
| 5.59e-248 | 1 | 427 | 3 | 425 | |
| 1.59e-247 | 1 | 427 | 3 | 425 | |
| 1.59e-247 | 1 | 427 | 3 | 425 | |
| 2.67e-247 | 1 | 427 | 17 | 439 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-74 | 6 | 391 | 31 | 425 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
|
| 4.32e-47 | 547 | 963 | 32 | 447 | Crystal structure of eukaryotic MATE transporter AtDTX14 [Arabidopsis thaliana] |
|
| 4.02e-34 | 515 | 952 | 36 | 468 | Chain A, Protein DETOXIFICATION [Nicotiana tabacum],7DQK_B Chain B, Protein DETOXIFICATION [Nicotiana tabacum] |
|
| 3.50e-33 | 525 | 939 | 13 | 426 | Crystal structure of a MATE family protein [Camelina sativa] |
|
| 3.77e-33 | 524 | 950 | 9 | 444 | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter [Vibrio cholerae O1 biovar El Tor str. N16961],3MKT_B Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter [Vibrio cholerae O1 biovar El Tor str. N16961],3MKU_A Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter [Vibrio cholerae O1 biovar El Tor str. N16961],3MKU_B Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter [Vibrio cholerae O1 biovar El Tor str. N16961] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.48e-169 | 1 | 427 | 22 | 445 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DFG5 PE=1 SV=1 |
|
| 4.68e-155 | 512 | 972 | 215 | 672 | Uncharacterized transporter YDR338C OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YDR338C PE=1 SV=1 |
|
| 1.24e-146 | 1 | 427 | 27 | 452 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
|
| 1.71e-128 | 3 | 427 | 21 | 445 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DCW1 PE=1 SV=1 |
|
| 4.58e-128 | 1 | 420 | 22 | 438 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=DCW1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000068 | 0.000001 |
TMHMM Annotations download full data without filtering help
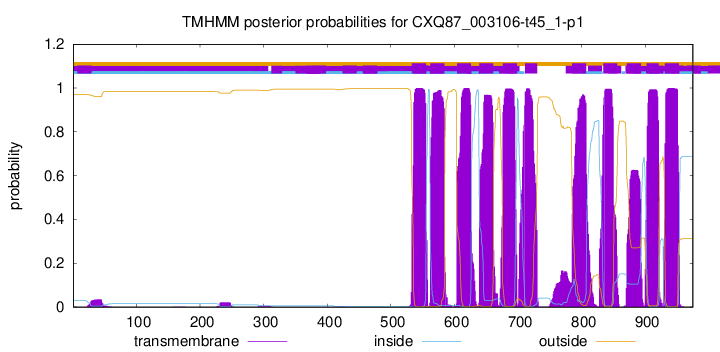
| Start | End |
|---|---|
| 533 | 555 |
| 562 | 584 |
| 604 | 626 |
| 639 | 661 |
| 676 | 698 |
| 711 | 730 |
| 785 | 807 |
| 834 | 856 |
| 871 | 893 |
| 902 | 921 |
| 931 | 953 |
