You are browsing environment: FUNGIDB
CAZyme Information: CXQ87_003026-t45_1-p1
You are here: Home > Sequence: CXQ87_003026-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] duobushaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] duobushaemulonis | |||||||||||
| CAZyme ID | CXQ87_003026-t45_1-p1 | |||||||||||
| CAZy Family | GT91 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 273337 | 2A060601 | 4.14e-149 | 39 | 725 | 286 | 1010 | Niemann-Pick C type protein family. The model describes Niemann-Pick C type protein in eukaryotes. The defective protein has been associated with Niemann-Pick disease which is described in humans as autosomal recessive lipidosis. It is characterized by the lysosomal accumulation of unestrified cholesterol. It is an integral membrane protein, which indicates that this protein is most likely involved in cholesterol transport or acts as some component of cholesterol homeostasis. [Transport and binding proteins, Other] |
| 403533 | Sterol-sensing | 2.74e-60 | 341 | 496 | 1 | 153 | Sterol-sensing domain of SREBP cleavage-activation. Sterol regulatory element-binding proteins (SREBPs) are membrane-bound transcription factors that promote lipid synthesis in animal cells. They are embedded in the membranes of the endoplasmic reticulum (ER) in a helical hairpin orientation and are released from the ER by a two-step proteolytic process. Proteolysis begins when the SREBPs are cleaved at Site-1, which is located at a leucine residue in the middle of the hydrophobic loop in the lumen of the ER. Upon proteolytic processing SREBP can activate the expression of genes involved in cholesterol biosynthesis and uptake. SCAP stimulates cleavage of SREBPs via fusion of the their two C-termini. This domain is the transmembrane region that traverses the membrane eight times and is the sterol-sensing domain of the cleavage protein. WD40 domains are found towards the C-terminus. |
| 308203 | Patched | 5.28e-37 | 241 | 679 | 150 | 571 | Patched family. The transmembrane protein Patched is a receptor for the morphogene Sonic Hedgehog. This protein associates with the smoothened protein to transduce hedgehog signals. |
| 273338 | 2A060602 | 1.86e-25 | 238 | 489 | 310 | 567 | The Eukaryotic (Putative) Sterol Transporter (EST) Family. |
| 199889 | E_set_AMPKbeta_like_N | 7.00e-23 | 745 | 818 | 7 | 79 | N-terminal Early set domain, a glycogen binding domain, associated with the catalytic domain of AMP-activated protein kinase beta subunit. E or "early" set domains are associated with the catalytic domain of AMP-activated protein kinase beta subunit glycogen binding domain at the N-terminal end. AMPK is a metabolic stress sensing protein that senses AMP/ATP and has recently been found to act as a glycogen sensor as well. The protein functions as an alpha-beta-gamma heterotrimer. This N-terminal domain is the glycogen binding domain of the beta subunit. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, and isoamylase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 3 | 1497 | 279 | 1913 | |
| 1.79e-130 | 747 | 1497 | 10 | 685 | |
| 3.45e-130 | 747 | 1497 | 10 | 685 | |
| 6.64e-130 | 747 | 1497 | 10 | 685 | |
| 2.21e-47 | 744 | 909 | 31 | 197 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.25e-163 | 63 | 734 | 323 | 972 | Crystal structure of S. cerevisia Niemann-Pick type C protein NCR1 [Saccharomyces cerevisiae S288C] |
|
| 2.35e-97 | 63 | 743 | 335 | 1070 | cryo-EM structure of the full-length human NPC1 at 4.4 angstrom [Homo sapiens],5JNX_A The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus [Homo sapiens] |
|
| 2.58e-97 | 63 | 743 | 335 | 1070 | Structure of itraconazole-bound NPC1 [Homo sapiens] |
|
| 3.17e-97 | 63 | 743 | 335 | 1070 | Crystal structure of human Niemann-Pick C1 protein [Homo sapiens],5U74_A Structure of human Niemann-Pick C1 protein [Homo sapiens] |
|
| 3.40e-97 | 63 | 743 | 335 | 1070 | NPC1 structure in Nanodisc [Homo sapiens],6W5S_A NPC1 structure in GDN micelles at pH 8.0 [Homo sapiens],6W5T_A NPC1 structure in GDN micelles at pH 5.5, conformation a [Homo sapiens],6W5U_A NPC1 structure in GDN micelles at pH 5.5, conformation b [Homo sapiens],6W5V_A NPC1-NPC2 complex structure at pH 5.5 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.28e-163 | 63 | 734 | 323 | 972 | NPC intracellular sterol transporter 1-related protein 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=NCR1 PE=1 SV=1 |
|
| 3.92e-99 | 62 | 747 | 334 | 1073 | NPC intracellular cholesterol transporter 1 OS=Sus scrofa OX=9823 GN=NPC1 PE=2 SV=1 |
|
| 1.21e-96 | 63 | 743 | 335 | 1070 | NPC intracellular cholesterol transporter 1 OS=Homo sapiens OX=9606 GN=NPC1 PE=1 SV=2 |
|
| 1.16e-93 | 63 | 747 | 335 | 1073 | NPC intracellular cholesterol transporter 1 OS=Mus musculus OX=10090 GN=Npc1 PE=1 SV=2 |
|
| 1.29e-91 | 39 | 777 | 309 | 1117 | NPC1-like intracellular cholesterol transporter 1 OS=Mus musculus OX=10090 GN=Npc1l1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000040 | 0.000008 |
TMHMM Annotations download full data without filtering help
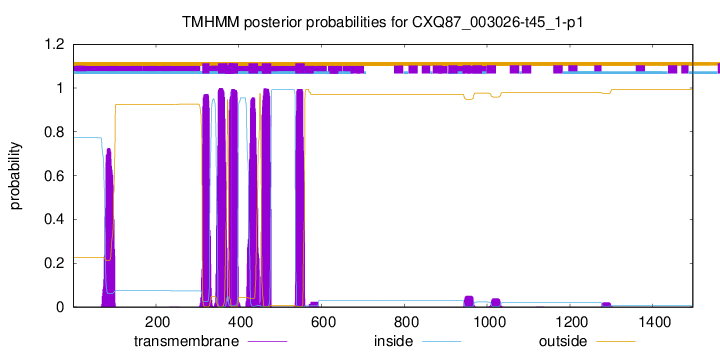
| Start | End |
|---|---|
| 312 | 329 |
| 350 | 372 |
| 376 | 398 |
| 424 | 446 |
| 456 | 478 |
| 537 | 559 |
