You are browsing environment: FUNGIDB
CAZyme Information: CXQ87_001272-t45_1-p1
You are here: Home > Sequence: CXQ87_001272-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] duobushaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] duobushaemulonis | |||||||||||
| CAZyme ID | CXQ87_001272-t45_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | adenylosuccinate synthetase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.259:12 | 2.4.1.261:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 475 | 716 | 2.7e-43 | 0.5784061696658098 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 240376 | PTZ00350 | 0.0 | 4 | 412 | 16 | 421 | adenylosuccinate synthetase; Provisional |
| 197875 | Adenylsucc_synt | 0.0 | 2 | 411 | 1 | 403 | Adenylosuccinate synthetase. Adenylosuccinate synthetase plays an important role in purine biosynthesis, by catalyzing the GTP-dependent conversion of IMP and aspartic acid to AMP. Adenylosuccinate synthetase has been characterized from various sources ranging from Escherichia coli (gene purA) to vertebrate tissues. In vertebrates, two isozymes are present - one involved in purine biosynthesis and the other in the purine nucleotide cycle. The crystal structure of adenylosuccinate synthetase from E. coli reveals that the dominant structural element of each monomer of the homodimer is a central beta-sheet of 10 strands. The first nine strands of the sheet are mutually parallel with right-handed crossover connections between the strands. The 10th strand is antiparallel with respect to the first nine strands. In addition, the enzyme has two antiparallel beta-sheets, comprised of two strands and three strands each, 11 alpha-helices and two short 3/10-helices. Further, it has been suggested that the similarities in the GTP-binding domains of the synthetase and the p21ras protein are an example of convergent evolution of two distinct families of GTP-binding proteins. Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana when compared with the known structures from E. coli reveals that the overall fold is very similar to that of the E. coli protein. |
| 234904 | PRK01117 | 0.0 | 3 | 412 | 4 | 410 | adenylosuccinate synthetase; Provisional |
| 223182 | PurA | 0.0 | 1 | 411 | 2 | 410 | Adenylosuccinate synthase [Nucleotide transport and metabolism]. |
| 272949 | purA | 0.0 | 4 | 411 | 1 | 405 | adenylosuccinate synthase. Alternate name IMP--aspartate ligase. [Purines, pyrimidines, nucleosides, and nucleotides, Purine ribonucleotide biosynthesis] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.00e-257 | 488 | 850 | 180 | 542 | |
| 7.24e-128 | 5 | 411 | 944 | 1348 | |
| 5.00e-119 | 488 | 850 | 216 | 578 | |
| 2.13e-102 | 494 | 849 | 217 | 582 | |
| 1.80e-99 | 501 | 847 | 247 | 596 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.13e-180 | 4 | 411 | 21 | 425 | Unligated adenylosuccinate synthetase from Cryptococcus neoformans [Cryptococcus neoformans var. grubii H99],5I33_B Unligated adenylosuccinate synthetase from Cryptococcus neoformans [Cryptococcus neoformans var. grubii H99],5I34_A Adenylosuccinate synthetase from Cryptococcus neoformans complexed with GDP and IMP [Cryptococcus neoformans var. grubii H99],5I34_B Adenylosuccinate synthetase from Cryptococcus neoformans complexed with GDP and IMP [Cryptococcus neoformans var. grubii H99] |
|
| 3.72e-157 | 4 | 411 | 35 | 443 | Human Adenylosuccinate synthetase isozyme 2 in complex with GDP [Homo sapiens] |
|
| 2.18e-155 | 4 | 411 | 35 | 441 | IMP Complex of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase [Mus musculus],1IWE_B IMP Complex of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase [Mus musculus],1J4B_A Recombinant Mouse-Muscle Adenylosuccinate Synthetase [Mus musculus],1LNY_A Crystal structure of the recombinant mouse-muscle adenylosuccinate synthetase complexed with 6-phosphoryl-IMP, GDP and Mg [Mus musculus],1LNY_B Crystal structure of the recombinant mouse-muscle adenylosuccinate synthetase complexed with 6-phosphoryl-IMP, GDP and Mg [Mus musculus],1LON_A Crystal Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with 6-phosphoryl-IMP, GDP and Hadacidin [Mus musculus],1LOO_A Crystal Structure of the Mouse-Muscle Adenylosuccinate Synthetase Ligated with GTP [Mus musculus],1MEZ_A Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) [Mus musculus],1MF0_A Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP, GDP, HPO4(2-), and Mg(2+) [Mus musculus],1MF1_A Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP [Mus musculus],2DGN_A Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP [Mus musculus] |
|
| 6.86e-136 | 5 | 411 | 23 | 427 | Structures Of Adenylosuccinate Synthetase From Triticum Aestivum And Arabidopsis Thaliana [Triticum aestivum],1DJ3_B Structures Of Adenylosuccinate Synthetase From Triticum Aestivum And Arabidopsis Thaliana [Triticum aestivum] |
|
| 1.89e-132 | 5 | 411 | 24 | 428 | Structures Of Adenylosuccinate Synthetase From Triticum Aestivum And Arabidopsis Thaliana [Arabidopsis thaliana],1DJ2_B Structures Of Adenylosuccinate Synthetase From Triticum Aestivum And Arabidopsis Thaliana [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.96e-255 | 1 | 411 | 1 | 411 | Adenylosuccinate synthetase OS=Candida albicans (strain WO-1) OX=294748 GN=ADE12 PE=3 SV=1 |
|
| 1.96e-255 | 1 | 411 | 1 | 411 | Adenylosuccinate synthetase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=ADE12 PE=3 SV=1 |
|
| 7.92e-255 | 1 | 411 | 1 | 411 | Adenylosuccinate synthetase OS=Candida dubliniensis (strain CD36 / ATCC MYA-646 / CBS 7987 / NCPF 3949 / NRRL Y-17841) OX=573826 GN=CD36_09080 PE=3 SV=1 |
|
| 3.69e-251 | 1 | 411 | 1 | 411 | Adenylosuccinate synthetase OS=Scheffersomyces stipitis (strain ATCC 58785 / CBS 6054 / NBRC 10063 / NRRL Y-11545) OX=322104 GN=PICST_85387 PE=3 SV=1 |
|
| 4.86e-251 | 1 | 411 | 1 | 411 | Adenylosuccinate synthetase OS=Candida tropicalis (strain ATCC MYA-3404 / T1) OX=294747 GN=CTRG_03394 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000013 | 0.000045 |
TMHMM Annotations download full data without filtering help
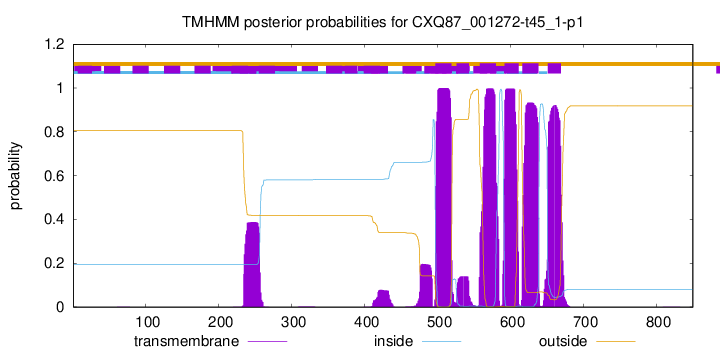
| Start | End |
|---|---|
| 497 | 519 |
| 526 | 543 |
| 558 | 580 |
| 592 | 611 |
| 616 | 638 |
| 651 | 669 |
