You are browsing environment: FUNGIDB
CAZyme Information: CXQ87_000678-t45_1-p1
You are here: Home > Sequence: CXQ87_000678-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] duobushaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] duobushaemulonis | |||||||||||
| CAZyme ID | CXQ87_000678-t45_1-p1 | |||||||||||
| CAZy Family | GH130 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.109:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 324 | 573 | 6.6e-64 | 0.9910313901345291 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224839 | PMT1 | 0.0 | 313 | 1022 | 19 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| 223461 | YHI9 | 9.69e-69 | 4 | 275 | 6 | 286 | Predicted epimerase YddE/YHI9, PhzF superfamily [General function prediction only]. |
| 406576 | PMT_4TMC | 7.19e-67 | 822 | 1018 | 1 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| 396786 | PMT | 1.32e-66 | 323 | 577 | 2 | 245 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This is a family of Dolichyl-phosphate-mannose-protein mannosyltransferase proteins EC:2.4.1.109. These proteins are responsible for O-linked glycosylation of proteins, they catalyze the reaction:- Dolichyl phosphate D-mannose + protein <=> dolichyl phosphate + O-D-mannosyl-protein. Also in this family is Drosophila rotated abdomen protein which is a putative mannosyltransferase. This family appears to be distantly related to pfam02516 (A Bateman pers. obs.). This family also contains sequences from ArnTs (4-amino-4-deoxy-L-arabinose lipid A transferase). They catalyze the addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the lipid A moiety of the lipopolysaccharide. This is a critical modification enabling bacteria (e.g. Escherichia coli and Salmonella typhimurium) to resist killing by antimicrobial peptides such as polymyxins. Members such as undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase are predicted to have 12 trans-membrane regions. The N-terminal portion of these proteins is hypothesized to have a conserved glycosylation activity which is shared between distantly related oligosaccharyltransferases ArnT and PglB families. |
| 129739 | PhzF_family | 5.41e-57 | 4 | 275 | 5 | 292 | phenazine biosynthesis protein PhzF family. Members of this family show a distant global similarity to diaminopimelate epimerases, which can be taken as the outgroup. One member of this family has been shown to act as an enzyme in the biosynthesis of the antibiotic phenazine in Pseudomonas aureofaciens. The function in other species is unclear. [Cellular processes, Toxin production and resistance] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1022 | 1 | 1025 | |
| 0.0 | 275 | 1022 | 26 | 780 | |
| 0.0 | 275 | 1022 | 26 | 780 | |
| 0.0 | 275 | 1022 | 26 | 780 | |
| 0.0 | 275 | 1022 | 26 | 780 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.68e-203 | 321 | 1022 | 68 | 753 | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor [Saccharomyces cerevisiae W303],6P2R_B Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor [Saccharomyces cerevisiae W303] |
|
| 1.34e-82 | 324 | 971 | 56 | 676 | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor [Saccharomyces cerevisiae W303],6P2R_A Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor [Saccharomyces cerevisiae W303] |
|
| 7.40e-47 | 4 | 275 | 7 | 289 | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily. [Saccharomyces cerevisiae] |
|
| 3.87e-45 | 602 | 803 | 11 | 205 | Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae],6ZQQ_B Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae],6ZQQ_C Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae],6ZQQ_D Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae] |
|
| 7.45e-42 | 602 | 803 | 2 | 195 | Crystal structure of the MIR domain (aa 337-532) of the S. cerevisiae mannosyltransferase Pmt2 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.25e-286 | 287 | 1022 | 45 | 832 | Dolichyl-phosphate-mannose--protein mannosyltransferase 6 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT6 PE=2 SV=1 |
|
| 4.17e-222 | 309 | 1022 | 57 | 757 | Dolichyl-phosphate-mannose--protein mannosyltransferase 6 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT6 PE=1 SV=1 |
|
| 1.60e-209 | 318 | 1022 | 66 | 763 | Dolichyl-phosphate-mannose--protein mannosyltransferase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT2 PE=1 SV=1 |
|
| 4.98e-202 | 321 | 1022 | 68 | 753 | Dolichyl-phosphate-mannose--protein mannosyltransferase 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT2 PE=1 SV=2 |
|
| 1.19e-191 | 286 | 1022 | 30 | 746 | Dolichyl-phosphate-mannose--protein mannosyltransferase 3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000000 |
TMHMM Annotations download full data without filtering help
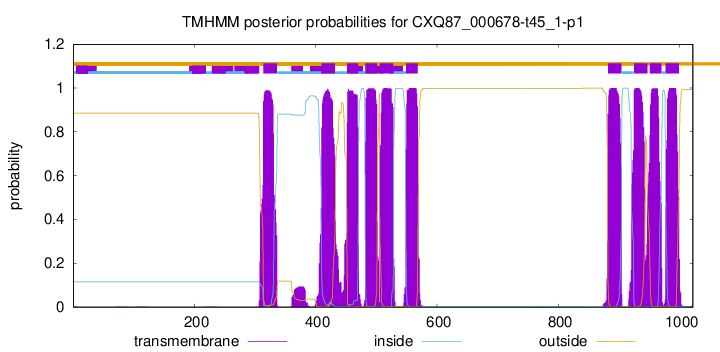
| Start | End |
|---|---|
| 314 | 336 |
| 410 | 432 |
| 452 | 470 |
| 482 | 501 |
| 506 | 528 |
| 549 | 568 |
| 882 | 904 |
| 925 | 947 |
| 951 | 970 |
| 977 | 999 |
