You are browsing environment: FUNGIDB
CAZyme Information: CXQ85_003418-t46_1-p1
You are here: Home > Sequence: CXQ85_003418-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] haemuloni | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] haemuloni | |||||||||||
| CAZyme ID | CXQ85_003418-t46_1-p1 | |||||||||||
| CAZy Family | GT3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT76 | 25 | 437 | 7.9e-89 | 0.9877149877149877 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 282094 | Mannosyl_trans2 | 5.83e-38 | 92 | 437 | 58 | 439 | Mannosyltransferase (PIG-V). This is a family of eukaryotic ER membrane proteins that are involved in the synthesis of glycosylphosphatidylinositol (GPI), a glycolipid that anchors many proteins to the eukaryotic cell surface. Proteins in this family are involved in transferring the second mannose in the biosynthetic pathway of GPI. |
| 227829 | COG5542 | 4.61e-17 | 143 | 437 | 121 | 408 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.07e-209 | 15 | 437 | 1 | 423 | |
| 1.00e-208 | 15 | 437 | 1 | 423 | |
| 1.10e-201 | 17 | 413 | 2 | 398 | |
| 2.13e-153 | 15 | 437 | 1 | 427 | |
| 4.28e-153 | 15 | 437 | 1 | 427 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.96e-115 | 29 | 437 | 15 | 428 | GPI mannosyltransferase 2 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI18 PE=3 SV=1 |
|
| 2.72e-86 | 20 | 437 | 4 | 394 | GPI mannosyltransferase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI18 PE=3 SV=1 |
|
| 1.90e-54 | 15 | 437 | 1 | 433 | GPI mannosyltransferase 2 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=GPI18 PE=3 SV=1 |
|
| 1.12e-50 | 90 | 437 | 56 | 417 | GPI mannosyltransferase 2 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=GPI18 PE=3 SV=1 |
|
| 3.81e-50 | 15 | 437 | 1 | 427 | GPI mannosyltransferase 2 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=GPI18 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000036 | 0.000005 |
TMHMM Annotations download full data without filtering help
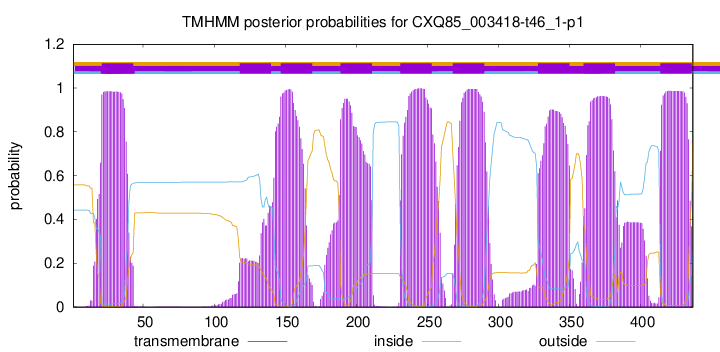
| Start | End |
|---|---|
| 21 | 43 |
| 118 | 140 |
| 147 | 169 |
| 189 | 211 |
| 231 | 253 |
| 268 | 290 |
| 328 | 350 |
| 360 | 382 |
| 414 | 436 |
