You are browsing environment: FUNGIDB
CAZyme Information: CXQ85_003045-t46_1-p1
You are here: Home > Sequence: CXQ85_003045-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] haemuloni | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] haemuloni | |||||||||||
| CAZyme ID | CXQ85_003045-t46_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 142 | 355 | 5.7e-68 | 0.7557251908396947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 206775 | RNase_PH_RRP41 | 1.04e-105 | 354 | 553 | 26 | 226 | RRP41 subunit of eukaryotic exosome. The RRP41 subunit of eukaryotic exosome is a member of the RNase_PH family, named after the bacterial Ribonuclease PH, a 3'-5' exoribonuclease. Structurally all members of this family form hexameric rings (trimers of Rrp41-Rrp45, Rrp46-Rrp43, and Mtr3-Rrp42 dimers). The eukaryotic exosome core is composed of six individually encoded RNase PH-like subunits and three additional proteins (Rrp4, Csl4 and Rrp40) that form a stable cap and contain RNA-binding domains. The RNase PH-like subunits are no longer phosphorolytic enzymes, the exosome directly associates with Rrp44 and Rrp6, hydrolytic exoribonucleases related to bacterial RNase II/R and RNase D. The exosome plays an important role in RNA turnover. It plays a crucial role in the maturation of stable RNA species such as rRNA, snRNA and snoRNA, quality control of mRNA, and the degradation of RNA processing by-products and non-coding transcripts. |
| 178434 | PLN02841 | 1.93e-66 | 7 | 353 | 6 | 350 | GPI mannosyltransferase |
| 252941 | Mannosyl_trans | 3.56e-58 | 142 | 353 | 1 | 194 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
| 223761 | Rph | 1.03e-52 | 353 | 549 | 31 | 230 | Ribonuclease PH [Translation, ribosomal structure and biogenesis]. |
| 235187 | PRK03983 | 5.97e-36 | 354 | 543 | 38 | 227 | exosome complex exonuclease Rrp41; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.39e-300 | 6 | 561 | 2 | 623 | |
| 3.64e-268 | 4 | 560 | 1 | 637 | |
| 3.49e-144 | 4 | 353 | 1 | 351 | |
| 5.63e-143 | 4 | 353 | 1 | 351 | |
| 2.26e-142 | 4 | 353 | 1 | 351 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.00e-75 | 354 | 555 | 37 | 239 | Cryo-EM structure of yeast cytoplasmic exosome [Saccharomyces cerevisiae],6LQS_R1 Chain R1, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C],7AJT_EC Chain EC, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C],7AJU_EC Chain EC, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C],7D4I_R1 Chain R1, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C] |
|
| 1.07e-75 | 354 | 555 | 39 | 241 | Crystal structure of an 11-subunit eukaryotic exosome complex bound to RNA [Saccharomyces cerevisiae S288C],5C0W_B Structure of a 12-subunit nuclear exosome complex bound to single-stranded RNA substrates [Saccharomyces cerevisiae S288C],5C0X_B Structure of a 12-subunit nuclear exosome complex bound to structured RNA [Saccharomyces cerevisiae S288C],6FSZ_BB Chain BB, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C] |
|
| 1.11e-75 | 354 | 555 | 40 | 242 | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA [Saccharomyces cerevisiae S288C],5OKZ_B Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C],5OKZ_L Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C],5OKZ_V Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C],5OKZ_f Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C] |
|
| 1.14e-75 | 354 | 555 | 41 | 243 | Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA [Saccharomyces cerevisiae S288C],5K36_B Structure of an eleven component nuclear RNA exosome complex bound to RNA [Saccharomyces cerevisiae S288C],5VZJ_B STRUCTURE OF A TWELVE COMPONENT MPP6-NUCLEAR RNA EXOSOME COMPLEX BOUND TO RNA [Saccharomyces cerevisiae S288C] |
|
| 1.99e-75 | 354 | 555 | 37 | 239 | yeast rrp44 nuclease [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.53e-106 | 4 | 354 | 1 | 339 | GPI mannosyltransferase 1 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI14 PE=3 SV=2 |
|
| 3.17e-101 | 16 | 354 | 11 | 328 | GPI mannosyltransferase 1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI14 PE=3 SV=1 |
|
| 5.17e-75 | 354 | 555 | 37 | 239 | Exosome complex component SKI6 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SKI6 PE=1 SV=1 |
|
| 8.19e-75 | 6 | 353 | 3 | 328 | GPI mannosyltransferase 1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=GPI14 PE=3 SV=1 |
|
| 5.71e-72 | 12 | 353 | 29 | 415 | GPI mannosyltransferase 1 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=gim-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000027 | 0.000005 |
TMHMM Annotations download full data without filtering help
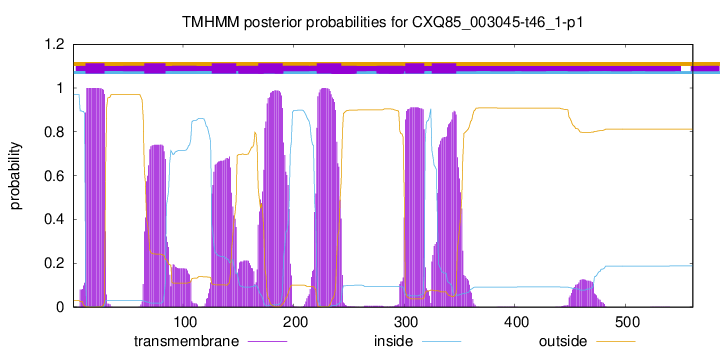
| Start | End |
|---|---|
| 12 | 29 |
| 65 | 84 |
| 126 | 148 |
| 168 | 190 |
| 221 | 243 |
| 301 | 318 |
| 325 | 347 |
