You are browsing environment: FUNGIDB
CAZyme Information: CXQ85_002361-t46_1-p1
You are here: Home > Sequence: CXQ85_002361-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] haemuloni | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] haemuloni | |||||||||||
| CAZyme ID | CXQ85_002361-t46_1-p1 | |||||||||||
| CAZy Family | GH72|CBM43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT91 | 653 | 1171 | 1.2e-123 | 0.975609756097561 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403385 | DUF3589 | 1.06e-175 | 643 | 1173 | 1 | 485 | Protein of unknown function (DUF3589). This family of proteins is found in eukaryotes. Proteins in this family are typically between 541 and 717 amino acids in length. The function of this family is not known, |
| 224374 | CodB | 1.98e-88 | 58 | 501 | 8 | 430 | Purine-cytosine permease or related protein [Nucleotide transport and metabolism]. |
| 271377 | SLC-NCS1sbd_CobB-like | 2.51e-65 | 62 | 508 | 1 | 405 | nucleobase-cation-symport-1 (NCS1) transporter CobB-like; solute-binding domain. This NCS1 subfamily includes Escherichia coli CodB (cytosine permease), and the Saccharomyces cerevisiae transporters: Fcy21p (Purine-cytosine permease), and vitamin B6 transporter Tpn1. NCS1s are essential components of salvage pathways for nucleobases and related metabolites; their known substrates include allantoin, uracil, thiamine, and nicotinamide riboside. NCS1s belong to a superfamily which also contains the solute carrier 5 family sodium/glucose transporters (SLC5s), and solute carrier 6 family neurotransmitter transporters (SLC6s). |
| 251109 | Transp_cyt_pur | 1.62e-55 | 62 | 489 | 2 | 436 | Permease for cytosine/purines, uracil, thiamine, allantoin. |
| 404414 | AA_permease_2 | 4.89e-06 | 85 | 461 | 5 | 402 | Amino acid permease. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1220 | 1 | 1152 | |
| 0.0 | 504 | 1222 | 1 | 719 | |
| 0.0 | 504 | 1222 | 1 | 719 | |
| 0.0 | 504 | 1222 | 1 | 719 | |
| 0.0 | 504 | 1222 | 1 | 719 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.76e-148 | 35 | 512 | 47 | 526 | Purine-cytosine permease FCY21 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FCY21 PE=1 SV=2 |
|
| 5.64e-144 | 43 | 512 | 58 | 531 | Purine-cytosine permease FCY2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FCY2 PE=1 SV=2 |
|
| 6.73e-139 | 43 | 512 | 55 | 528 | Purine-cytosine permease FCY22 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FCY22 PE=3 SV=1 |
|
| 2.31e-112 | 630 | 1207 | 102 | 678 | Beta-mannosyltransferase 8 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BMT8 PE=3 SV=1 |
|
| 5.40e-101 | 45 | 512 | 43 | 506 | Purine-cytosine permease fcyB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=fcyB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000070 | 0.000001 |
TMHMM Annotations download full data without filtering help
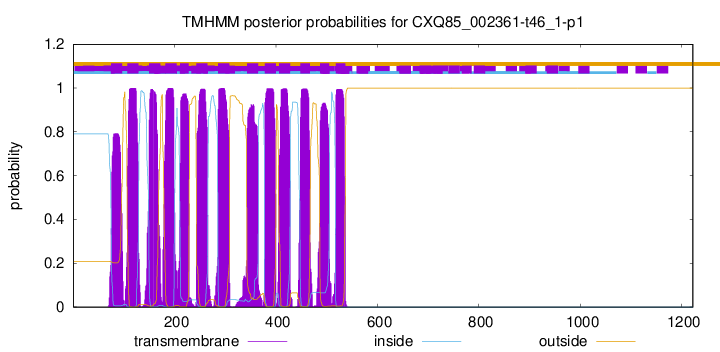
| Start | End |
|---|---|
| 75 | 97 |
| 107 | 129 |
| 150 | 172 |
| 182 | 204 |
| 211 | 228 |
| 243 | 265 |
| 286 | 308 |
| 343 | 365 |
| 378 | 400 |
| 406 | 428 |
| 448 | 467 |
| 487 | 505 |
| 518 | 537 |
