You are browsing environment: FUNGIDB
CAZyme Information: CXQ85_001914-t46_1-p1
You are here: Home > Sequence: CXQ85_001914-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] haemuloni | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] haemuloni | |||||||||||
| CAZyme ID | CXQ85_001914-t46_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.131:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340835 | GT4_ALG11-like | 0.0 | 97 | 500 | 1 | 390 | alpha-1,2-mannosyltransferase ALG11 and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. ALG11 in yeast is involved in adding the final 1,2-linked Man to the Man5GlcNAc2-PP-Dol synthesized on the cytosolic face of the ER. The deletion analysis of ALG11 was shown to block the early steps of core biosynthesis that takes place on the cytoplasmic face of the ER and lead to a defect in the assembly of lipid-linked oligosaccharides. |
| 397941 | Coatomer_WDAD | 9.43e-153 | 902 | 1405 | 1 | 439 | Coatomer WD associated region. This region is composed of WD40 repeats. |
| 406369 | ALG11_N | 3.46e-98 | 97 | 306 | 1 | 208 | ALG11 mannosyltransferase N-terminus. |
| 215511 | PLN02949 | 1.07e-89 | 93 | 468 | 30 | 393 | transferase, transferring glycosyl groups |
| 238121 | WD40 | 2.04e-64 | 599 | 872 | 11 | 279 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.48e-314 | 1 | 580 | 1 | 592 | |
| 5.86e-312 | 1 | 580 | 1 | 592 | |
| 1.66e-311 | 1 | 580 | 1 | 592 | |
| 2.65e-310 | 1 | 580 | 1 | 592 | |
| 3.96e-265 | 1 | 576 | 1 | 598 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-235 | 585 | 1430 | 1 | 802 | Crystal structure of yeast alpha/betaprime-COP subcomplex of the COPI vesicular coat [Saccharomyces cerevisiae YJM789],3MKQ_C Crystal structure of yeast alpha/betaprime-COP subcomplex of the COPI vesicular coat [Saccharomyces cerevisiae YJM789],3MKQ_E Crystal structure of yeast alpha/betaprime-COP subcomplex of the COPI vesicular coat [Saccharomyces cerevisiae YJM789] |
|
| 7.70e-194 | 585 | 1421 | 3 | 777 | The structure of the COPI coat triad [Mus musculus],5A1V_D The structure of the COPI coat linkage I [Mus musculus],5A1V_L The structure of the COPI coat linkage I [Mus musculus],5A1V_U The structure of the COPI coat linkage I [Mus musculus],5A1W_D The structure of the COPI coat linkage II [Mus musculus],5A1X_D The structure of the COPI coat linkage III [Mus musculus],5A1X_L The structure of the COPI coat linkage III [Mus musculus],5A1Y_D The structure of the COPI coat linkage IV [Mus musculus],5A1Y_L The structure of the COPI coat linkage IV [Mus musculus],5NZR_C The structure of the COPI coat leaf [Mus musculus],5NZS_C The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor [Mus musculus],5NZT_C The structure of the COPI coat linkage I [Mus musculus],5NZT_H The structure of the COPI coat linkage I [Mus musculus],5NZU_C The structure of the COPI coat linkage II [Mus musculus],5NZV_C The structure of the COPI coat linkage IV [Mus musculus],5NZV_J The structure of the COPI coat linkage IV [Mus musculus] |
|
| 1.66e-189 | 585 | 1201 | 1 | 604 | yeast betaprime COP 1-604 with KTKTN motif [Saccharomyces cerevisiae] |
|
| 1.48e-96 | 585 | 886 | 1 | 304 | yeast betaprime COP 1-304 with KTKTN motif [Saccharomyces cerevisiae] |
|
| 1.81e-96 | 585 | 886 | 1 | 304 | yeast betaprime COP 1-304H6 [Saccharomyces cerevisiae],2YNO_B yeast betaprime COP 1-304H6 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.35e-233 | 585 | 1430 | 1 | 802 | Coatomer subunit beta' OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SEC27 PE=1 SV=1 |
|
| 3.98e-200 | 585 | 1441 | 3 | 793 | Coatomer subunit beta'-1 OS=Arabidopsis thaliana OX=3702 GN=At1g79990 PE=2 SV=2 |
|
| 5.61e-197 | 8 | 580 | 8 | 602 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=ALG11 PE=3 SV=2 |
|
| 1.52e-195 | 16 | 580 | 19 | 609 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=ALG11 PE=3 SV=2 |
|
| 9.64e-195 | 585 | 1421 | 3 | 777 | Coatomer subunit beta' OS=Rattus norvegicus OX=10116 GN=Copb2 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
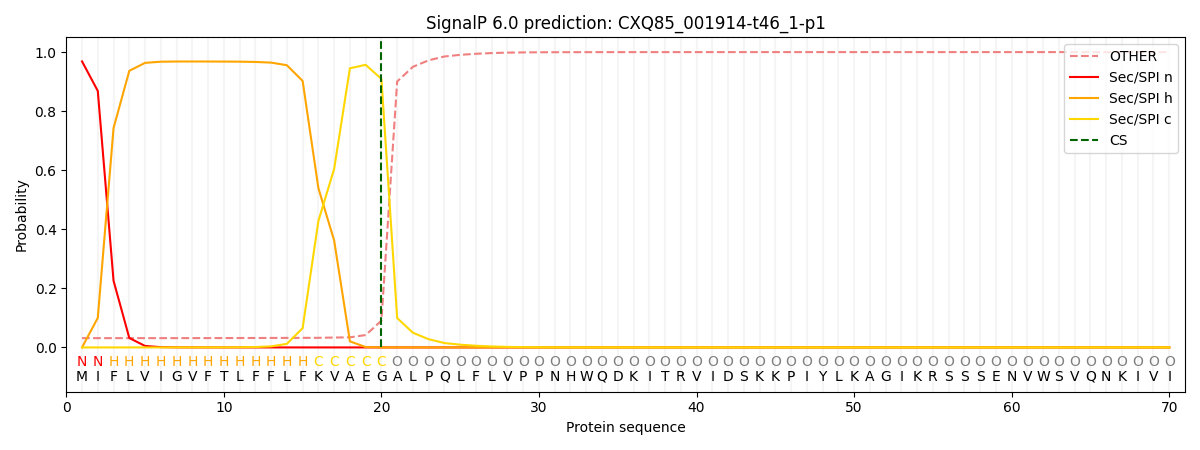
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.037072 | 0.962911 | CS pos: 20-21. Pr: 0.9094 |
