You are browsing environment: FUNGIDB
CAZyme Information: CXQ85_001892-t46_1-p1
You are here: Home > Sequence: CXQ85_001892-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] haemuloni | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] haemuloni | |||||||||||
| CAZyme ID | CXQ85_001892-t46_1-p1 | |||||||||||
| CAZy Family | GH38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 102 | 273 | 3.1e-78 | 0.9885057471264368 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397941 | Coatomer_WDAD | 8.00e-158 | 808 | 1239 | 1 | 439 | Coatomer WD associated region. This region is composed of WD40 repeats. |
| 185692 | GH16_fungal_CRH1_transglycosylase | 8.08e-95 | 90 | 298 | 2 | 203 | glycosylphosphatidylinositol-glucanosyltransferase. Group of fungal GH16 members related to Saccharomyces cerevisiae Crh1p. Chr1p and Crh2p are transglycosylases that are required for the linkage of chitin to beta(1-3)glucose branches of beta(1-6)glucan, an important step in the assembly of new cell wall. Both have been shown to be glycosylphosphatidylinositol (GPI)-anchored. A third homologous protein, Crr1p, functions in the formation of the spore wall. They belongs to the family 16 of glycosyl hydrolases that includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| 399736 | COPI_C | 1.50e-80 | 1379 | 1674 | 104 | 403 | Coatomer (COPI) alpha subunit C-terminus. This family represents the C-terminus (approximately 500 residues) of the eukaryotic coatomer alpha subunit. Coatomer (COPI) is a large cytosolic protein complex which forms a coat around vesicles budding from the Golgi apparatus. Such coatomer-coated vesicles have been proposed to play a role in many distinct steps of intracellular transport. Note that many family members also contain the pfam04053 domain. |
| 238121 | WD40 | 2.39e-79 | 478 | 787 | 15 | 289 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
| 395585 | Glyco_hydro_16 | 1.24e-55 | 103 | 269 | 3 | 168 | Glycosyl hydrolases family 16. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.57e-222 | 1 | 451 | 1 | 449 | |
| 2.56e-221 | 1 | 451 | 1 | 449 | |
| 3.60e-221 | 1 | 451 | 1 | 449 | |
| 3.60e-221 | 1 | 451 | 1 | 449 | |
| 4.38e-204 | 1 | 451 | 2439 | 2895 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.13e-241 | 464 | 1674 | 1 | 1218 | The structure of the COPI coat triad [Mus musculus],5A1V_C The structure of the COPI coat linkage I [Mus musculus],5A1V_K The structure of the COPI coat linkage I [Mus musculus],5A1V_T The structure of the COPI coat linkage I [Mus musculus],5A1W_C The structure of the COPI coat linkage II [Mus musculus],5A1X_C The structure of the COPI coat linkage III [Mus musculus],5A1X_K The structure of the COPI coat linkage III [Mus musculus],5A1Y_C The structure of the COPI coat linkage IV [Mus musculus],5A1Y_K The structure of the COPI coat linkage IV [Mus musculus],5NZR_A The structure of the COPI coat leaf [Mus musculus],5NZS_A The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor [Mus musculus],5NZT_A The structure of the COPI coat linkage I [Mus musculus],5NZU_A The structure of the COPI coat linkage II [Mus musculus],5NZV_A The structure of the COPI coat linkage IV [Mus musculus],5NZV_H The structure of the COPI coat linkage IV [Mus musculus] |
|
| 9.05e-88 | 1376 | 1676 | 2 | 303 | Crystal structure of yeast alpha/epsilon-COP of the COPI vesicular coat [Saccharomyces cerevisiae S288C] |
|
| 1.36e-87 | 1371 | 1676 | 19 | 325 | Crystal Structure of a-COP in Complex with e-COP [Saccharomyces cerevisiae],3MV2_C Crystal Structure of a-COP in Complex with e-COP [Saccharomyces cerevisiae],3MV2_E Crystal Structure of a-COP in Complex with e-COP [Saccharomyces cerevisiae] |
|
| 1.62e-86 | 1371 | 1676 | 19 | 325 | Crystal Structure of a-COP in Complex with e-COP [Saccharomyces cerevisiae],3MV3_C Crystal Structure of a-COP in Complex with e-COP [Saccharomyces cerevisiae],3MV3_E Crystal Structure of a-COP in Complex with e-COP [Saccharomyces cerevisiae] |
|
| 1.36e-61 | 462 | 789 | 1 | 327 | Crystal structure of alpha-COP [Schizosaccharomyces pombe],4J8B_A Crystal structure of alpha-COP/Emp47p complex [Schizosaccharomyces pombe],4J8G_A Crystal structure of alpha-COP/E19 complex [Schizosaccharomyces pombe],4J8G_B Crystal structure of alpha-COP/E19 complex [Schizosaccharomyces pombe] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 462 | 1676 | 1 | 1201 | Coatomer subunit alpha OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=COP1 PE=1 SV=2 |
|
| 3.39e-287 | 462 | 1673 | 1 | 1204 | Putative coatomer subunit alpha OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBPJ4664.04 PE=1 SV=1 |
|
| 1.39e-241 | 464 | 1674 | 1 | 1218 | Coatomer subunit alpha OS=Homo sapiens OX=9606 GN=COPA PE=1 SV=2 |
|
| 1.39e-241 | 464 | 1674 | 1 | 1218 | Coatomer subunit alpha OS=Bos taurus OX=9913 GN=COPA PE=1 SV=1 |
|
| 1.94e-241 | 464 | 1674 | 1 | 1218 | Coatomer subunit alpha OS=Mus musculus OX=10090 GN=Copa PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
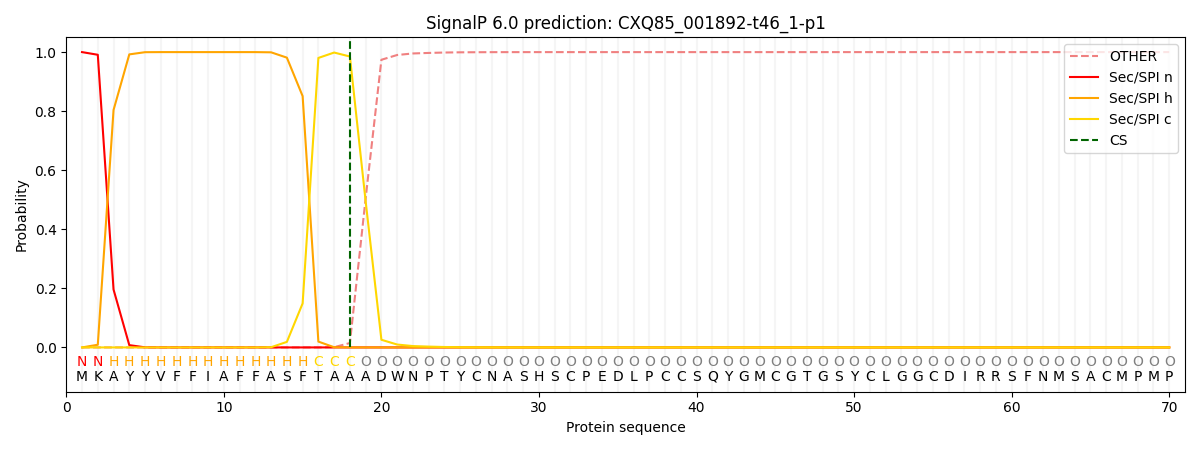
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000214 | 0.999762 | CS pos: 18-19. Pr: 0.9851 |
