You are browsing environment: FUNGIDB
CAZyme Information: CVL01589.1
You are here: Home > Sequence: CVL01589.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium mangiferae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium mangiferae | |||||||||||
| CAZyme ID | CVL01589.1 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | Glycosidase [Source:UniProtKB/TrEMBL;Acc:A0A1L7TUB1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 105 | 268 | 5.4e-79 | 0.9425287356321839 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185692 | GH16_fungal_CRH1_transglycosylase | 8.38e-86 | 85 | 293 | 2 | 203 | glycosylphosphatidylinositol-glucanosyltransferase. Group of fungal GH16 members related to Saccharomyces cerevisiae Crh1p. Chr1p and Crh2p are transglycosylases that are required for the linkage of chitin to beta(1-3)glucose branches of beta(1-6)glucan, an important step in the assembly of new cell wall. Both have been shown to be glycosylphosphatidylinositol (GPI)-anchored. A third homologous protein, Crr1p, functions in the formation of the spore wall. They belongs to the family 16 of glycosyl hydrolases that includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| 395585 | Glyco_hydro_16 | 2.86e-56 | 99 | 264 | 3 | 168 | Glycosyl hydrolases family 16. |
| 225182 | BglS | 7.26e-23 | 128 | 315 | 117 | 291 | Beta-glucanase, GH16 family [Carbohydrate transport and metabolism]. |
| 211314 | ChtBD1_GH16 | 2.04e-22 | 19 | 65 | 1 | 47 | Hevein or Type 1 chitin binding domain subfamily that co-occurs with family 16 glycosyl hydrolases. This subfamily includes Saccharomyces cerevisiae Utr2p, also known as Crh2p, which participates in the cross-linking of chitin to beta(1-3)- and beta(1-6) glucan in the cell wall, and S. cerevisiae Crr1p, a putative transglycosidase which is needed for proper spore wall assembly. ChtBD1 is a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 185685 | GH16_XET | 5.52e-22 | 108 | 311 | 28 | 224 | Xyloglucan endotransglycosylase, member of glycosyl hydrolase family 16. Xyloglucan endotransglycosylases (XETs) cleave and religate xyloglucan polymers in plant cell walls via a transglycosylation mechanism. Xyloglucan is a soluble hemicellulose with a backbone of beta-1,4-linked glucose units, partially substituted with alpha-1,6-linked xylopyranose branches. It binds noncovalently to cellulose, cross-linking the adjacent cellulose microfibrils, giving it a key structural role as a matrix polymer. Therefore, XET plays an important role in all plant processes that require cell wall remodeling. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.78e-304 | 1 | 475 | 1 | 477 | |
| 1.78e-304 | 1 | 475 | 1 | 477 | |
| 1.78e-304 | 1 | 475 | 1 | 477 | |
| 1.78e-304 | 1 | 475 | 1 | 477 | |
| 1.00e-276 | 1 | 475 | 1 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.63e-32 | 96 | 318 | 37 | 242 | Apo Crh5 transglycosylase [Aspergillus fumigatus Af293],6IBU_B Apo Crh5 transglycosylase [Aspergillus fumigatus Af293],6IBW_A Crh5 transglycosylase in complex with NAG [Aspergillus fumigatus Af293],6IBW_B Crh5 transglycosylase in complex with NAG [Aspergillus fumigatus Af293] |
|
| 1.93e-15 | 92 | 250 | 22 | 188 | Chain A, Beta-glucanase [Acetivibrio thermocellus ATCC 27405],3WVJ_B Chain B, Beta-glucanase [Acetivibrio thermocellus ATCC 27405] |
|
| 3.75e-13 | 155 | 274 | 87 | 193 | Crystal Structure of the catalytic nucleophile and surface cysteine mutant of VvEG16 in complex with a xyloglucan oligosaccharide [Vitis vinifera] |
|
| 1.69e-12 | 155 | 274 | 86 | 192 | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with cellotetraose [Vitis vinifera] |
|
| 1.72e-12 | 155 | 274 | 87 | 193 | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with a mixed-linkage glucan octasaccharide [Vitis vinifera],5DZF_B Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with a mixed-linkage glucan octasaccharide [Vitis vinifera] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.31e-133 | 9 | 340 | 11 | 349 | Probable glycosidase crf2 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=crf2 PE=1 SV=1 |
|
| 9.43e-109 | 21 | 336 | 29 | 354 | Extracellular glycosidase UTR2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=UTR2 PE=1 SV=1 |
|
| 1.22e-105 | 17 | 319 | 23 | 336 | Probable glycosidase CRH2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=UTR2 PE=1 SV=3 |
|
| 4.83e-67 | 19 | 295 | 29 | 357 | Probable glycosidase CRR1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CRR1 PE=2 SV=1 |
|
| 3.84e-66 | 19 | 307 | 29 | 377 | Probable glycosidase CRR1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=CRR1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
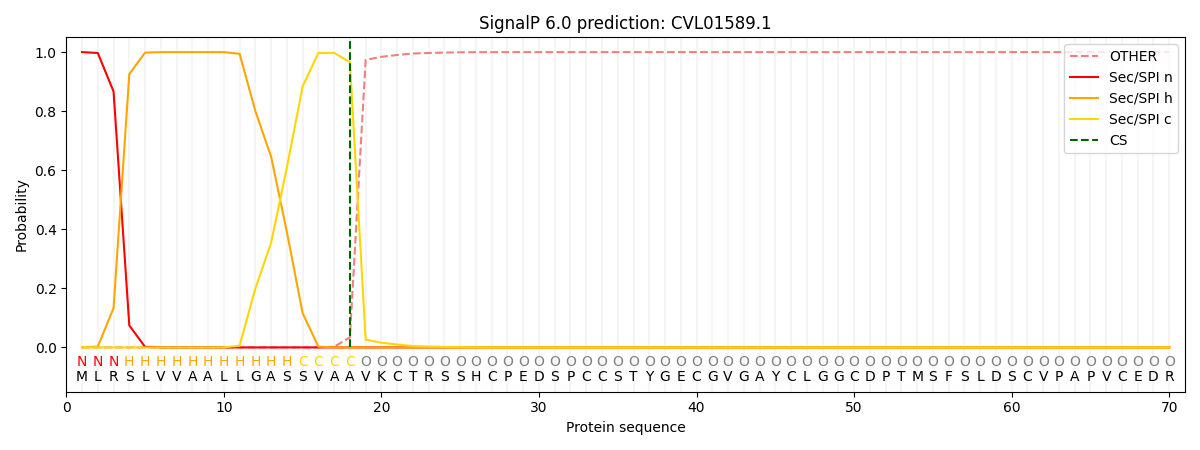
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000314 | 0.999659 | CS pos: 18-19. Pr: 0.9659 |
