You are browsing environment: FUNGIDB
CAZyme Information: CVL00961.1
You are here: Home > Sequence: CVL00961.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
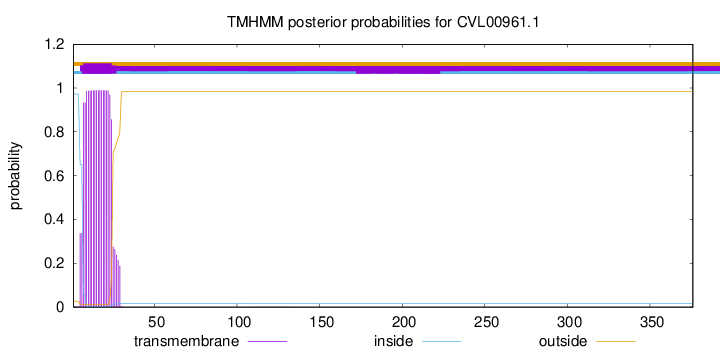
TMHMM annotations
Basic Information help
| Species | Fusarium mangiferae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium mangiferae | |||||||||||
| CAZyme ID | CVL00961.1 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | Related to beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A1L7U0C6] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT17 | 67 | 364 | 4.6e-82 | 0.9859154929577465 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398414 | Glyco_transf_17 | 2.29e-51 | 60 | 363 | 50 | 342 | Glycosyltransferase family 17. This family represents beta-1,4-mannosyl-glycoprotein beta-1,4-N-acetylglucosaminyltransferase (EC:2.4.1.144). This enzyme transfers the bisecting GlcNAc to the core mannose of complex N-glycans. The addition of this residue is regulated during development and has functional consequences for receptor signalling, cell adhesion, and tumor progression. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.45e-286 | 1 | 376 | 1 | 376 | |
| 3.07e-284 | 1 | 376 | 1 | 376 | |
| 3.07e-284 | 1 | 376 | 1 | 376 | |
| 3.07e-284 | 1 | 376 | 1 | 376 | |
| 3.07e-284 | 1 | 376 | 1 | 376 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.39e-10 | 89 | 364 | 206 | 499 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Homo sapiens OX=9606 GN=MGAT3 PE=1 SV=3 |
|
| 4.42e-10 | 89 | 364 | 210 | 503 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Rattus norvegicus OX=10116 GN=Mgat3 PE=1 SV=2 |
|
| 5.88e-10 | 89 | 364 | 210 | 503 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Mus musculus OX=10090 GN=Mgat3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
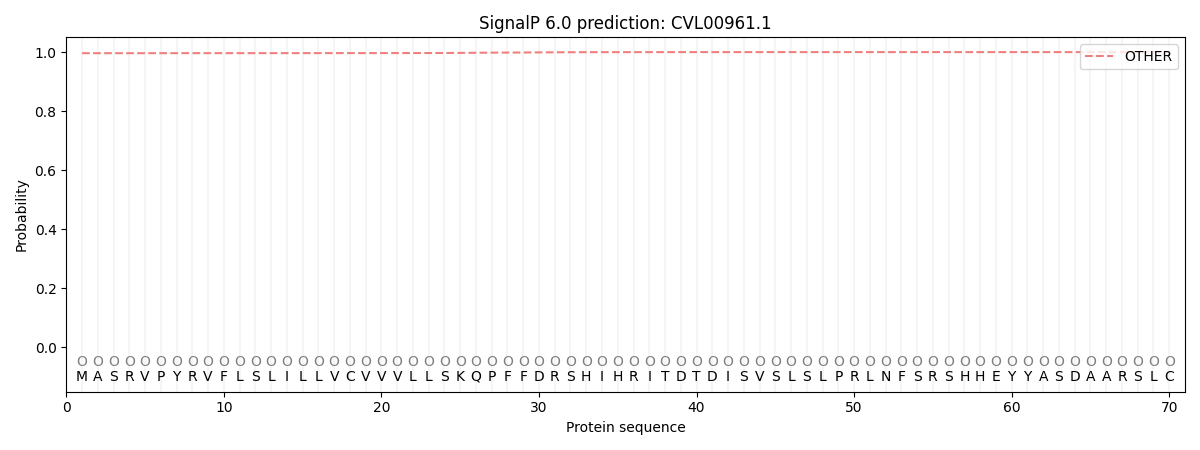
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.996477 | 0.003548 |