You are browsing environment: FUNGIDB
CAZyme Information: CTRG_02349-t43_1-p1
You are here: Home > Sequence: CTRG_02349-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida tropicalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida tropicalis | |||||||||||
| CAZyme ID | CTRG_02349-t43_1-p1 | |||||||||||
| CAZy Family | GH65 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.256:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 35 | 393 | 6.8e-105 | 0.995049504950495 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398537 | DIE2_ALG10 | 5.07e-127 | 36 | 393 | 1 | 383 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.56e-166 | 1 | 425 | 1 | 437 | |
| 1.33e-154 | 29 | 425 | 2 | 420 | |
| 1.37e-145 | 1 | 425 | 1 | 444 | |
| 1.18e-141 | 1 | 425 | 1 | 444 | |
| 2.75e-121 | 1 | 425 | 1 | 453 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.42e-161 | 1 | 425 | 1 | 450 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DIE2 PE=3 SV=1 |
|
| 8.82e-105 | 19 | 425 | 10 | 449 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=ALG10 PE=3 SV=1 |
|
| 8.89e-70 | 34 | 425 | 64 | 514 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DIE2 PE=1 SV=2 |
|
| 1.69e-69 | 36 | 425 | 57 | 498 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=ALG10 PE=3 SV=2 |
|
| 3.65e-67 | 33 | 425 | 60 | 511 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=ALG10 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000018 | 0.000005 |
TMHMM Annotations download full data without filtering help
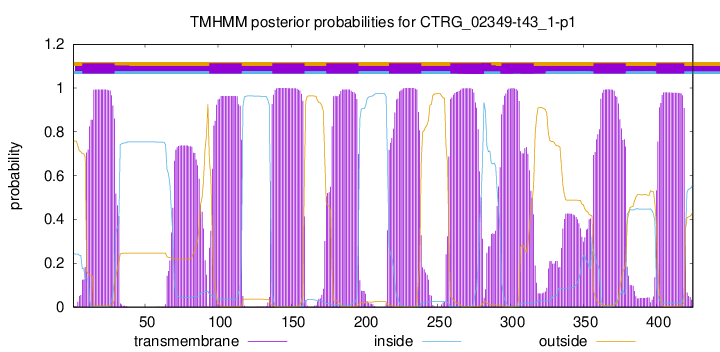
| Start | End |
|---|---|
| 7 | 29 |
| 94 | 116 |
| 137 | 159 |
| 174 | 196 |
| 217 | 239 |
| 259 | 281 |
| 294 | 316 |
| 357 | 379 |
| 400 | 419 |
