You are browsing environment: FUNGIDB
CAZyme Information: CTRG_02093-t43_1-p1
You are here: Home > Sequence: CTRG_02093-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida tropicalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida tropicalis | |||||||||||
| CAZyme ID | CTRG_02093-t43_1-p1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 118 | 369 | 1e-23 | 0.9581993569131833 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.81e-97 | 97 | 370 | 28 | 303 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 2.03e-06 | 178 | 370 | 96 | 299 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.94e-176 | 1 | 370 | 1 | 373 | |
| 1.26e-169 | 1 | 370 | 32 | 409 | |
| 1.95e-166 | 1 | 370 | 1 | 379 | |
| 2.52e-164 | 1 | 370 | 1 | 378 | |
| 1.17e-152 | 1 | 370 | 1 | 376 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.48e-165 | 1 | 370 | 1 | 378 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
|
| 3.17e-126 | 114 | 370 | 134 | 388 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 4.27e-118 | 114 | 370 | 131 | 385 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
|
| 6.67e-47 | 114 | 370 | 306 | 563 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 6.67e-47 | 114 | 370 | 306 | 563 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
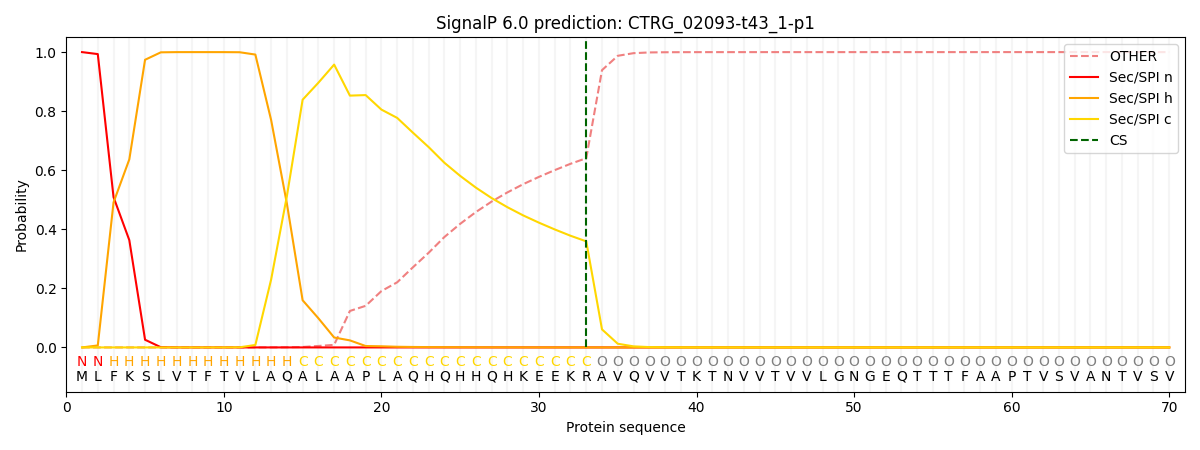
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999715 | CS pos: 33-34. Pr: 0.3592 |
