You are browsing environment: FUNGIDB
CAZyme Information: CTRG_01318-t43_1-p1
You are here: Home > Sequence: CTRG_01318-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida tropicalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida tropicalis | |||||||||||
| CAZyme ID | CTRG_01318-t43_1-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 12 | 406 | 5.2e-76 | 0.987146529562982 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 1.20e-49 | 10 | 405 | 1 | 410 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 215437 | PLN02816 | 1.89e-08 | 9 | 385 | 39 | 423 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.17e-277 | 1 | 502 | 9 | 506 | |
| 2.04e-276 | 1 | 502 | 1 | 498 | |
| 4.14e-222 | 7 | 502 | 39 | 518 | |
| 2.62e-212 | 7 | 502 | 6 | 485 | |
| 5.58e-200 | 6 | 502 | 5 | 507 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.62e-277 | 1 | 502 | 1 | 498 | GPI mannosyltransferase 4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=SMP3 PE=3 SV=2 |
|
| 3.27e-196 | 2 | 502 | 3 | 535 | GPI mannosyltransferase 4 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=SMP3 PE=3 SV=2 |
|
| 8.56e-95 | 23 | 501 | 19 | 513 | GPI mannosyltransferase 4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SMP3 PE=1 SV=3 |
|
| 2.20e-88 | 23 | 501 | 18 | 501 | GPI mannosyltransferase 4 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=SMP3 PE=3 SV=2 |
|
| 4.22e-86 | 15 | 502 | 12 | 510 | GPI mannosyltransferase 4 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=SMP3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999919 | 0.000112 |
TMHMM Annotations download full data without filtering help
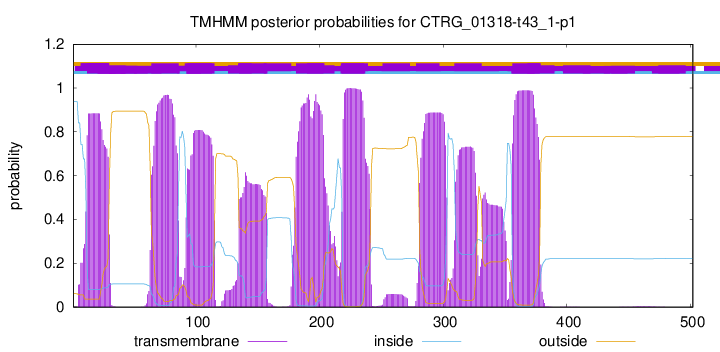
| Start | End |
|---|---|
| 12 | 29 |
| 64 | 86 |
| 93 | 115 |
| 135 | 157 |
| 181 | 203 |
| 218 | 236 |
| 356 | 378 |
