You are browsing environment: FUNGIDB
CAZyme Information: CPC735_057160-t26_1-p1
You are here: Home > Sequence: CPC735_057160-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
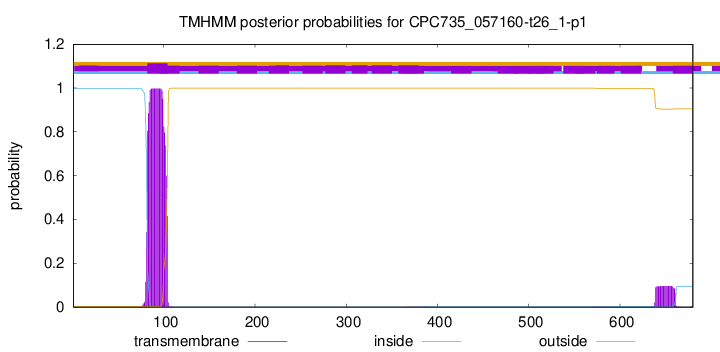
TMHMM annotations
Basic Information help
| Species | Coccidioides posadasii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Onygenaceae; Coccidioides; Coccidioides posadasii | |||||||||||
| CAZyme ID | CPC735_057160-t26_1-p1 | |||||||||||
| CAZy Family | GT21 | |||||||||||
| CAZyme Description | Multicopper oxidase family protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 155 | 655 | 9e-95 | 0.9804469273743017 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 3.12e-70 | 141 | 659 | 4 | 522 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 177843 | PLN02191 | 9.19e-66 | 137 | 658 | 22 | 544 | L-ascorbate oxidase |
| 215324 | PLN02604 | 2.82e-64 | 143 | 667 | 29 | 553 | oxidoreductase |
| 259977 | CuRO_3_MCO_like_4 | 7.49e-63 | 499 | 660 | 1 | 166 | The third cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidize their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This subfamily of MCOs is composed of three cupredoxin domains. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 274556 | laccase | 4.14e-57 | 162 | 667 | 27 | 526 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.16e-317 | 269 | 680 | 1 | 412 | |
| 1.07e-209 | 207 | 679 | 1 | 483 | |
| 3.12e-209 | 87 | 660 | 28 | 598 | |
| 7.92e-209 | 87 | 660 | 45 | 615 | |
| 7.92e-209 | 87 | 660 | 45 | 615 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.32e-57 | 138 | 658 | 2 | 477 | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_B Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_C Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_D Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_E Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_F Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae] |
|
| 3.46e-54 | 152 | 656 | 81 | 538 | Structure of the L499M mutant of the laccase from B.aclada [Botrytis aclada] |
|
| 9.02e-54 | 152 | 656 | 81 | 538 | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution [Botrytis aclada],4X4K_A Structure of laccase from Botrytis aclada with full copper content [Botrytis aclada] |
|
| 1.44e-49 | 144 | 658 | 9 | 467 | Native fungus laccase from Trametes hirsuta [Trametes hirsuta],3V9C_A Type-2 Cu-depleted fungus laccase from Trametes hirsuta at low dose of ionization radiation [Trametes hirsuta] |
|
| 3.73e-49 | 144 | 658 | 9 | 467 | Type-2 Cu-depleted fungus laccase from Trametes hirsuta [Trametes hirsuta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.78e-66 | 152 | 654 | 38 | 490 | Iron transport multicopper oxidase FET3 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FET3 PE=2 SV=1 |
|
| 6.83e-66 | 124 | 675 | 47 | 573 | Laccase-1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=LAC1 PE=1 SV=1 |
|
| 5.02e-64 | 124 | 663 | 47 | 563 | Laccase-2 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=LAC2 PE=3 SV=2 |
|
| 3.33e-63 | 124 | 667 | 47 | 567 | Laccase-1 OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=LAC1 PE=1 SV=1 |
|
| 2.05e-62 | 138 | 677 | 22 | 508 | Multicopper oxidase PfmaD OS=Pestalotiopsis fici (strain W106-1 / CGMCC3.15140) OX=1229662 GN=PfmaD PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
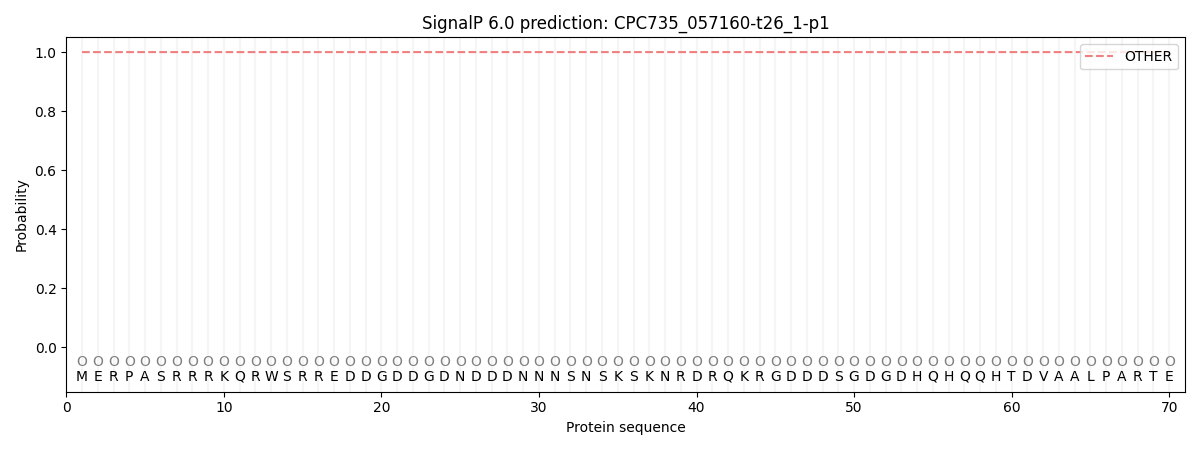
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000035 | 0.000000 |