You are browsing environment: FUNGIDB
CAZyme Information: CPAR2_213690-T-p1
You are here: Home > Sequence: CPAR2_213690-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida parapsilosis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida parapsilosis | |||||||||||
| CAZyme ID | CPAR2_213690-T-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | Has domain(s) with predicted catalytic activity and role in carbohydrate metabolic process | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 175 | 408 | 1.9e-28 | 0.6536312849162011 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 4.41e-10 | 212 | 437 | 35 | 291 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 398697 | LANC_like | 1.92e-05 | 231 | 359 | 130 | 267 | Lanthionine synthetase C-like protein. Lanthionines are thioether bridges that are putatively generated by dehydration of Ser and Thr residues followed by addition of cysteine residues within the peptide. This family contains the lanthionine synthetase C-like proteins 1 and 2 which are related to the bacterial lanthionine synthetase components C (LanC). LANCL1 (P40 seven-transmembrane-domain protein) and LANCL2 (testes-specific adriamycin sensitivity protein) are thought to be peptide-modifying enzyme components in eukaryotic cells. Both proteins are produced in large quantities in the brain and testes and may have role in the immune surveillance of these organs. Lanthionines are found in lantibiotics, which are peptide-derived, post-translationally modified antimicrobials produced by several bacterial strains. This region contains seven internal repeats. |
| 271198 | LanC_like | 2.27e-04 | 235 | 372 | 31 | 155 | Cyclases involved in the biosynthesis of lantibiotics, and similar proteins. LanC is the cyclase enzyme of the lanthionine synthetase. Lanthionine is a lantibiotic, a unique class of peptide antibiotics. They are ribosomally synthesized as a precursor peptide and then post-translationally modified to contain thioether cross-links called lanthionines (Lans) or methyllanthionines (MeLans), in addition to 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb). These unusual amino acids are introduced by the dehydration of serine and threonine residues, followed by thioether formation via addition of cysteine thiols, catalysed by LanB and LanC or LanM. LanC, the cyclase component, is a zinc metalloprotein, whose bound metal has been proposed to activate the thiol substrate for nucleophilic addition. A related domain is also present in LanM and other pro- and eukaryotic proteins with poorly characterized functions. |
| 271198 | LanC_like | 0.002 | 267 | 351 | 105 | 192 | Cyclases involved in the biosynthesis of lantibiotics, and similar proteins. LanC is the cyclase enzyme of the lanthionine synthetase. Lanthionine is a lantibiotic, a unique class of peptide antibiotics. They are ribosomally synthesized as a precursor peptide and then post-translationally modified to contain thioether cross-links called lanthionines (Lans) or methyllanthionines (MeLans), in addition to 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb). These unusual amino acids are introduced by the dehydration of serine and threonine residues, followed by thioether formation via addition of cysteine thiols, catalysed by LanB and LanC or LanM. LanC, the cyclase component, is a zinc metalloprotein, whose bound metal has been proposed to activate the thiol substrate for nucleophilic addition. A related domain is also present in LanM and other pro- and eukaryotic proteins with poorly characterized functions. |
| 224250 | YyaL | 0.008 | 263 | 304 | 274 | 315 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 532 | 1 | 532 | |
| 1.59e-293 | 1 | 532 | 1 | 538 | |
| 1.96e-134 | 172 | 532 | 85 | 447 | |
| 2.85e-101 | 176 | 530 | 53 | 416 | |
| 1.51e-89 | 177 | 527 | 212 | 571 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.55e-07 | 248 | 403 | 94 | 265 | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 9.83e-06 | 260 | 434 | 116 | 300 | Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
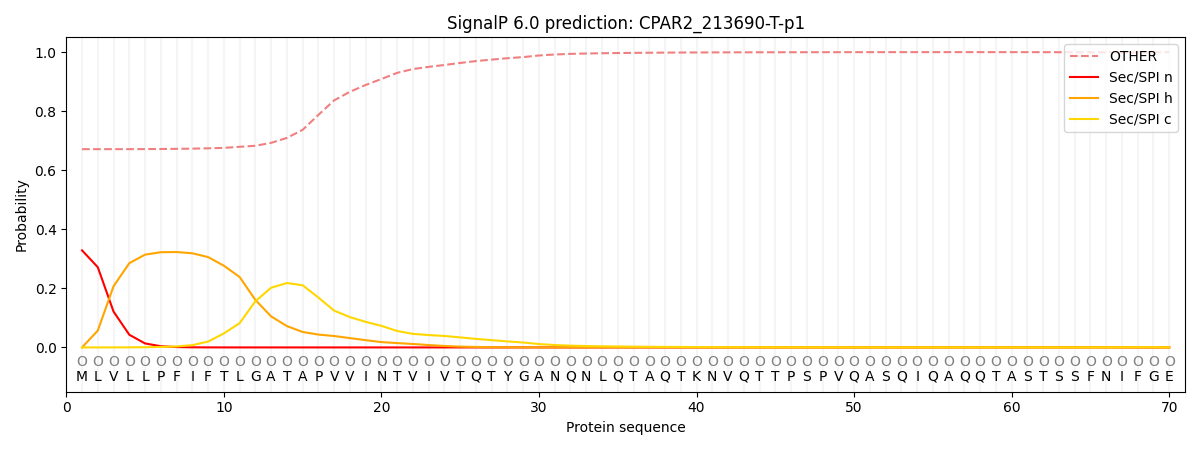
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.685508 | 0.314493 |
