You are browsing environment: FUNGIDB
CAZyme Information: CPAG_08635-t26_1-p1
Basic Information
help
| Species |
Coccidioides posadasii
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Onygenaceae; Coccidioides; Coccidioides posadasii
|
| CAZyme ID |
CPAG_08635-t26_1-p1
|
| CAZy Family |
GT41 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 374 |
|
40576.34 |
7.0582 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_CposadasiiRMSCC3488 |
10016 |
454284 |
119 |
9897
|
|
| Gene Location |
No EC number prediction in CPAG_08635-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA11 |
98 |
216 |
1.5e-34 |
0.6178010471204188 |
CPAG_08635-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.81e-289 |
1 |
374 |
14 |
387 |
| 3.62e-95 |
50 |
364 |
8 |
293 |
| 7.39e-95 |
50 |
364 |
8 |
283 |
| 1.38e-92 |
49 |
364 |
7 |
294 |
| 2.00e-78 |
54 |
358 |
12 |
309 |
CPAG_08635-t26_1-p1 has no PDB hit.
CPAG_08635-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
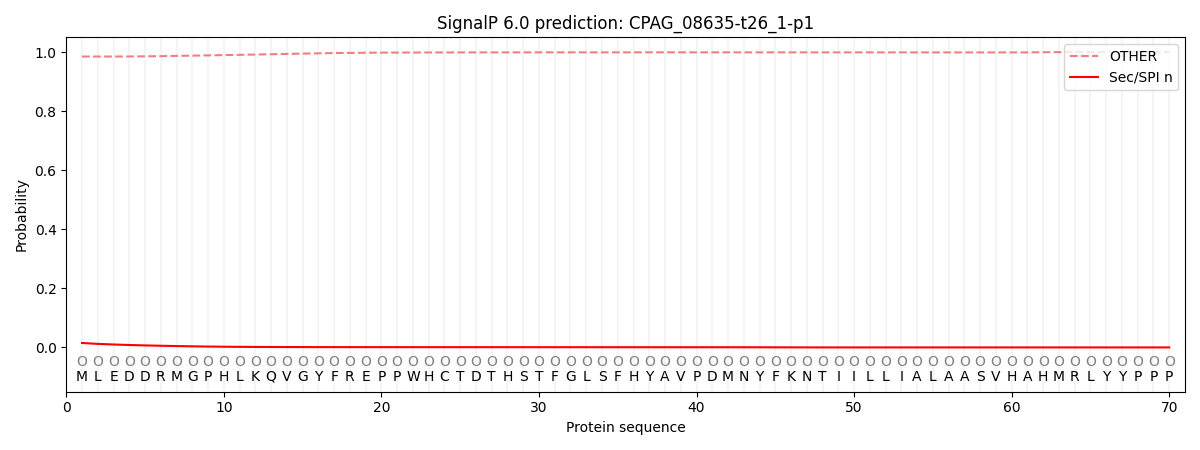
| Other |
SP_Sec_SPI |
CS Position |
| 0.986510 |
0.013477 |
|
There is no transmembrane helices in CPAG_08635-t26_1-p1.

