You are browsing environment: FUNGIDB
CNL06720-t26_1-p1
Basic Information
help
Species
Cryptococcus neoformans
Lineage
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans
CAZyme ID
CNL06720-t26_1-p1
CAZy Family
GT48
CAZyme Description
expressed protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
422
AE017352.1|CGC5 45092.94
4.0807
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CneoformansJEC21
7004
214684
372
6632
Gene Location
No EC number prediction in CNL06720-t26_1-p1.
Family
Start
End
Evalue
family coverage
GH105
139
353
3.9e-30
0.6566265060240963
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
400034
Glyco_hydro_88
1.95e-12
144
248
96
204
Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases.
more
226678
YesR
5.67e-11
144
320
102
284
Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
2.19e-305
1
422
1
422
4.95e-286
1
422
1
422
1.16e-284
1
422
1
422
1.16e-284
1
422
1
422
1.16e-284
1
422
1
422
CNL06720-t26_1-p1 has no PDB hit.
CNL06720-t26_1-p1 has no Swissprot hit.
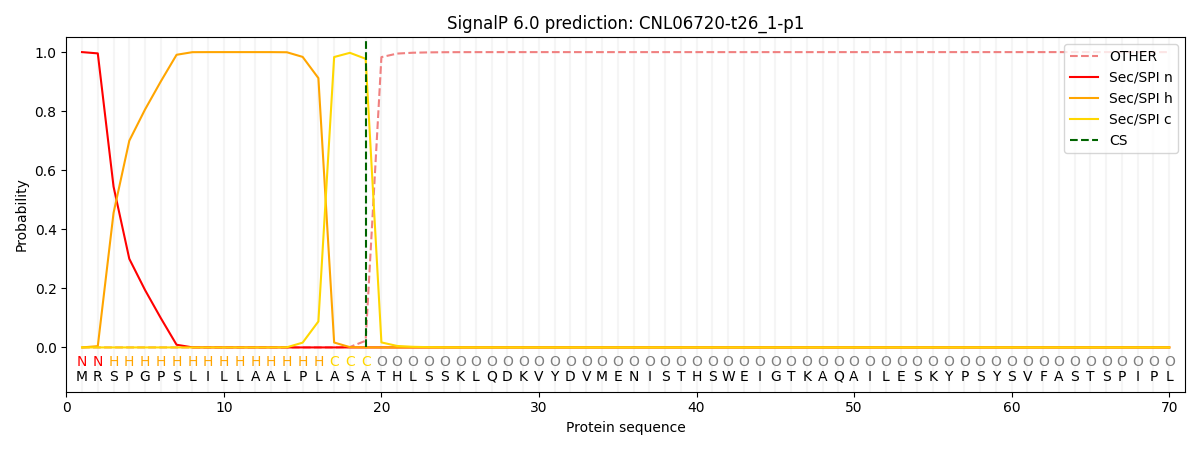
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000228
0.999738
CS pos: 19-20. Pr: 0.9774
There is no transmembrane helices in CNL06720-t26_1-p1.