You are browsing environment: FUNGIDB
CAZyme Information: CNG01530-t26_1-p1
You are here: Home > Sequence: CNG01530-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cryptococcus neoformans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans | |||||||||||
| CAZyme ID | CNG01530-t26_1-p1 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | endo alpha-1,4 polygalactosaminidase precusor, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH114 | 118 | 326 | 1.6e-64 | 0.9368421052631579 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397552 | Glyco_hydro_114 | 6.70e-70 | 96 | 326 | 1 | 191 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| 226386 | COG3868 | 1.49e-16 | 120 | 326 | 79 | 266 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.73e-240 | 1 | 359 | 1 | 359 | |
| 5.83e-240 | 1 | 359 | 13 | 371 | |
| 2.84e-214 | 1 | 359 | 1 | 358 | |
| 7.77e-212 | 1 | 359 | 1 | 358 | |
| 7.77e-212 | 1 | 359 | 1 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.05e-27 | 119 | 330 | 55 | 238 | Crystal Structure of Aspergillus fumigatus Ega3 [Aspergillus fumigatus Af293] |
|
| 8.45e-27 | 119 | 330 | 72 | 255 | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine [Aspergillus fumigatus Af293] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
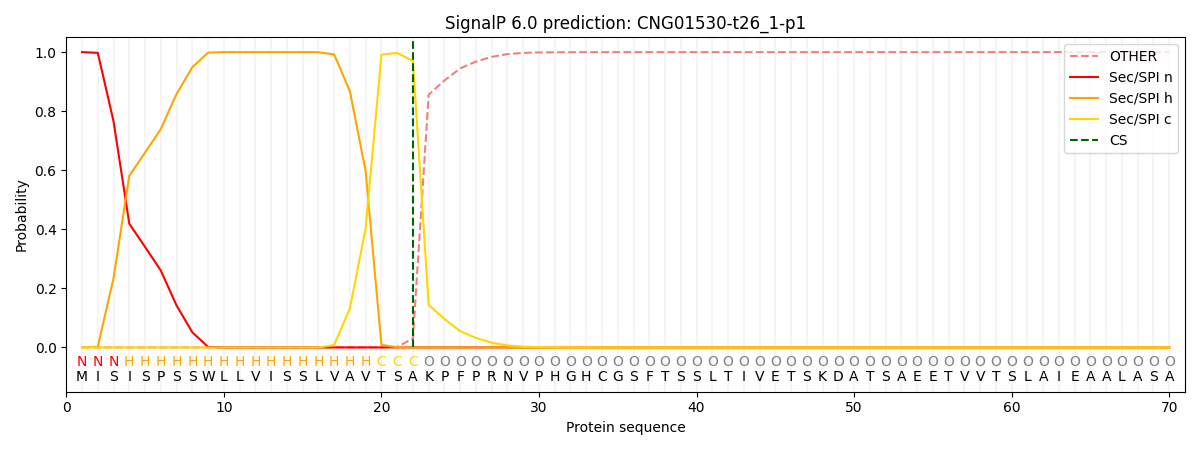
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000258 | 0.999707 | CS pos: 22-23. Pr: 0.9696 |
