You are browsing environment: FUNGIDB
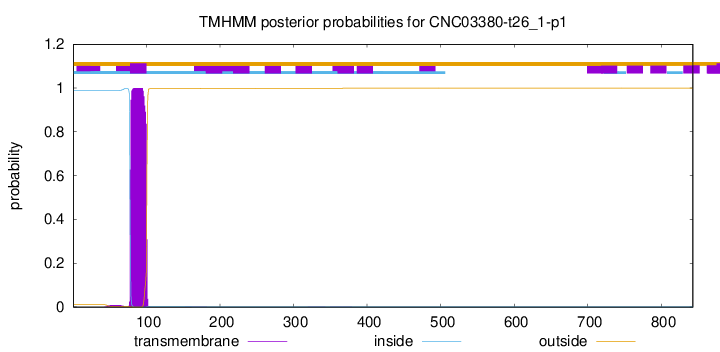
CNC03380-t26_1-p1
Basic Information
help
Species
Cryptococcus neoformans
Lineage
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans
CAZyme ID
CNC03380-t26_1-p1
CAZy Family
CE9
CAZyme Description
expressed protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
843
AE017343.1|CGC1 90963.07
4.1761
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CneoformansJEC21
7004
214684
372
6632
Gene Location
No EC number prediction in CNC03380-t26_1-p1.
Family
Start
End
Evalue
family coverage
PL35
581
762
6.5e-46
0.9720670391061452
CNC03380-t26_1-p1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
0.0
1
843
1
843
0.0
1
843
1
841
0.0
1
843
1
841
0.0
1
843
1
836
0.0
1
843
1
842
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5.55e-06
269
736
206
661
Crystal structure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58],3A0O_B Crystal structure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58]
more
CNC03380-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000043
0.000000