You are browsing environment: FUNGIDB
CNBG3070-t26_1-p1
Basic Information
help
Species
Cryptococcus neoformans
Lineage
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans
CAZyme ID
CNBG3070-t26_1-p1
CAZy Family
GH5
CAZyme Description
unspecified product
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
411
42812.42
5.3326
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CneoformansB-3501A
6608
283643
108
6500
Gene Location
No EC number prediction in CNBG3070-t26_1-p1.
Family
Start
End
Evalue
family coverage
PL14
153
364
5.5e-73
0.9857142857142858
CNBG3070-t26_1-p1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.19e-269
1
411
1
401
2.33e-245
1
411
1
396
2.33e-245
1
411
1
396
5.46e-244
1
411
1
396
1.84e-235
1
411
1
396
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.87e-41
154
364
58
270
Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai]
more
1.28e-09
193
364
74
237
Structure of alginate lyase Aly36B [Chitinophaga sp. MD30]
more
CNBG3070-t26_1-p1 has no Swissprot hit.
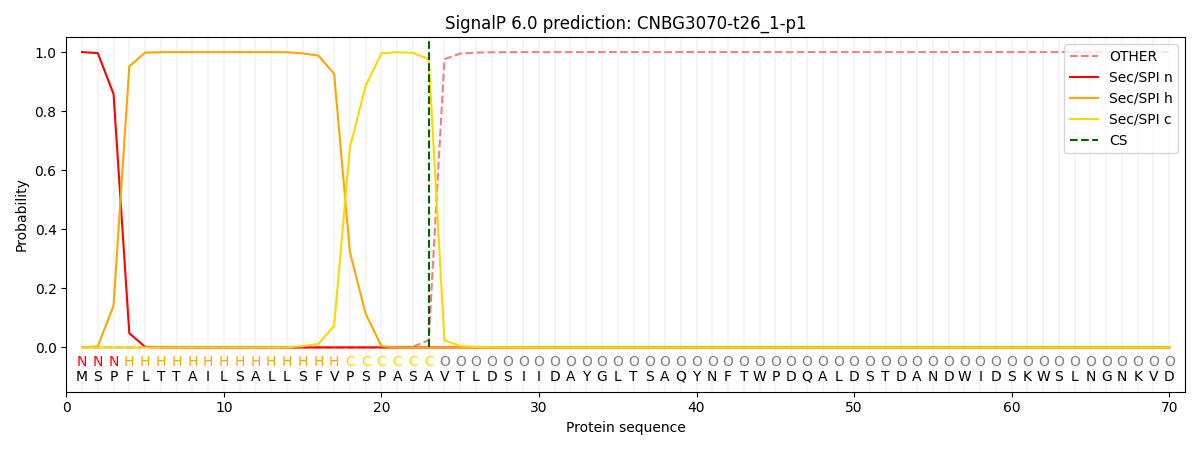
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000226
0.999746
CS pos: 23-24. Pr: 0.9758
There is no transmembrane helices in CNBG3070-t26_1-p1.