You are browsing environment: FUNGIDB
CAZyme Information: CNBC1160-t26_1-p1
You are here: Home > Sequence: CNBC1160-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cryptococcus neoformans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans | |||||||||||
| CAZyme ID | CNBC1160-t26_1-p1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 199889 | E_set_AMPKbeta_like_N | 5.22e-21 | 8 | 83 | 3 | 77 | N-terminal Early set domain, a glycogen binding domain, associated with the catalytic domain of AMP-activated protein kinase beta subunit. E or "early" set domains are associated with the catalytic domain of AMP-activated protein kinase beta subunit glycogen binding domain at the N-terminal end. AMPK is a metabolic stress sensing protein that senses AMP/ATP and has recently been found to act as a glycogen sensor as well. The protein functions as an alpha-beta-gamma heterotrimer. This N-terminal domain is the glycogen binding domain of the beta subunit. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, and isoamylase. |
| 199892 | E_set_Isoamylase_like_N | 2.07e-16 | 8 | 83 | 4 | 83 | N-terminal Early set domain associated with the catalytic domain of isoamylase-like (also called glycogen 6-glucanohydrolase) proteins. E or "early" set domains are associated with the catalytic domain of isoamylase-like proteins at the N-terminal end. Isoamylase is one of the starch-debranching enzymes that catalyze the hydrolysis of alpha-1,6-glucosidic linkages specific in alpha-glucans such as amylopectin or glycogen. Isoamylase contains a bound calcium ion, but this is not in the same position as the conserved calcium ion that has been reported in other alpha-amylase family enzymes. The N-terminal domain of isoamylase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, and the beta subunit of AMP-activated protein kinase. |
| 406865 | AMPK1_CBM | 2.95e-13 | 8 | 85 | 4 | 78 | Glycogen recognition site of AMP-activated protein kinase. AMPK1_CBM is a family found in close association with AMPKBI pfam04739. The surface of AMPK1_CBM reveals a carbohydrate-binding pocket. |
| 199885 | E_set_GBE_prok_N | 8.43e-05 | 8 | 64 | 22 | 82 | N-terminal Early set domain associated with the catalytic domain of prokaryotic glycogen branching enzyme. This subfamily is composed of predominantly prokaryotic 1,4 alpha glucan branching enzymes, also called glycogen branching enzymes. E or "early" set domains are associated with the catalytic domain of glycogen branching enzymes at the N-terminal end. Glycogen branching enzyme catalyzes the formation of alpha-1,6 branch points in either glycogen or starch by cleavage of the alpha-1,4 glucosidic linkage, yielding a non-reducing end oligosaccharide chain, as well as the subsequent attachment of short glucosyl chains to the alpha-1,6 position. By increasing the number of non-reducing ends, glycogen is more reactive to synthesis and digestion as well as being more soluble. The N-terminal domain of the 1,4 alpha glucan branching enzyme may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| 223021 | PHA03247 | 7.82e-04 | 130 | 344 | 2751 | 2963 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 793 | 1 | 765 | |
| 0.0 | 1 | 793 | 1 | 740 | |
| 0.0 | 1 | 782 | 1 | 755 | |
| 0.0 | 1 | 793 | 1 | 741 | |
| 0.0 | 1 | 793 | 1 | 741 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.85e-06 | 8 | 82 | 10 | 86 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain YJM789) OX=307796 GN=CRP1 PE=3 SV=1 |
|
| 8.85e-06 | 8 | 82 | 10 | 86 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain RM11-1a) OX=285006 GN=CRP1 PE=3 SV=1 |
|
| 8.85e-06 | 8 | 82 | 10 | 86 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain JAY291) OX=574961 GN=CRP1 PE=3 SV=1 |
|
| 8.85e-06 | 8 | 82 | 10 | 86 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain Lalvin EC1118 / Prise de mousse) OX=643680 GN=CRP1 PE=3 SV=1 |
|
| 8.85e-06 | 8 | 82 | 10 | 86 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain AWRI796) OX=764097 GN=CRP1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
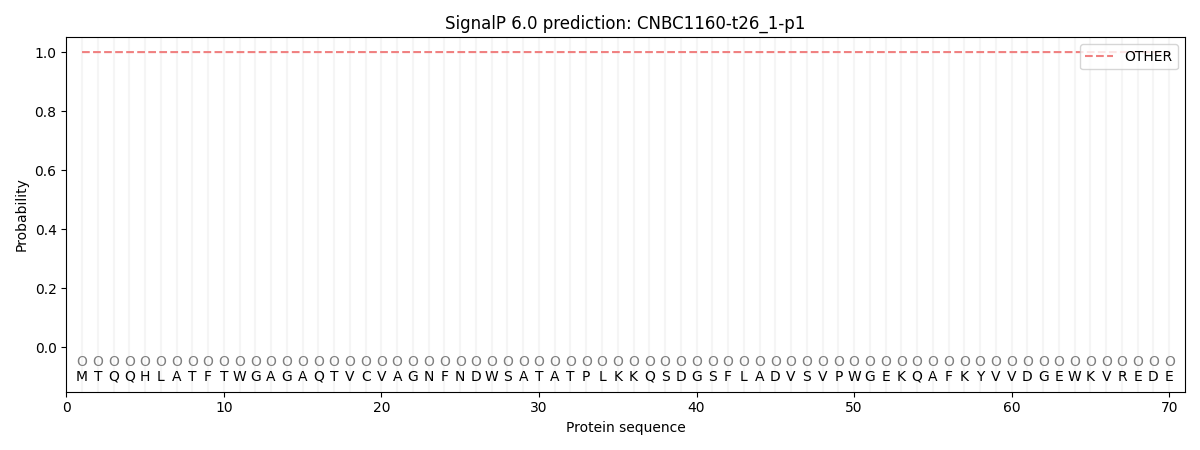
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000063 | 0.000000 |
