You are browsing environment: FUNGIDB
CAZyme Information: CNBA1510-t26_1-p1
You are here: Home > Sequence: CNBA1510-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
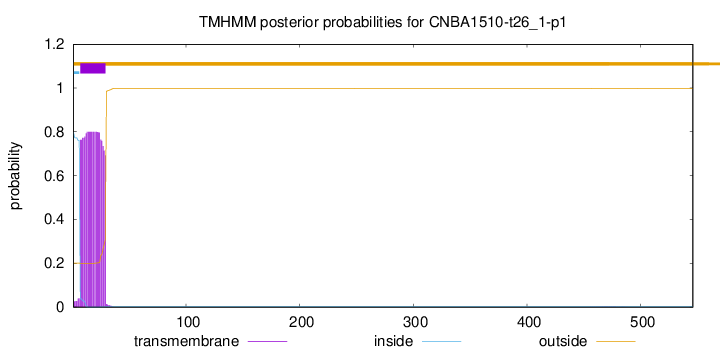
TMHMM annotations
Basic Information help
| Species | Cryptococcus neoformans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans | |||||||||||
| CAZyme ID | CNBA1510-t26_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA14 | 25 | 282 | 6.5e-114 | 0.9882352941176471 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.90e-281 | 1 | 546 | 1 | 546 | |
| 1.88e-224 | 6 | 546 | 1 | 541 | |
| 1.61e-217 | 6 | 545 | 1 | 527 | |
| 8.11e-214 | 1 | 545 | 1 | 531 | |
| 2.97e-211 | 6 | 546 | 1 | 541 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.89e-80 | 25 | 281 | 1 | 261 | Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina],5NO7_B Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
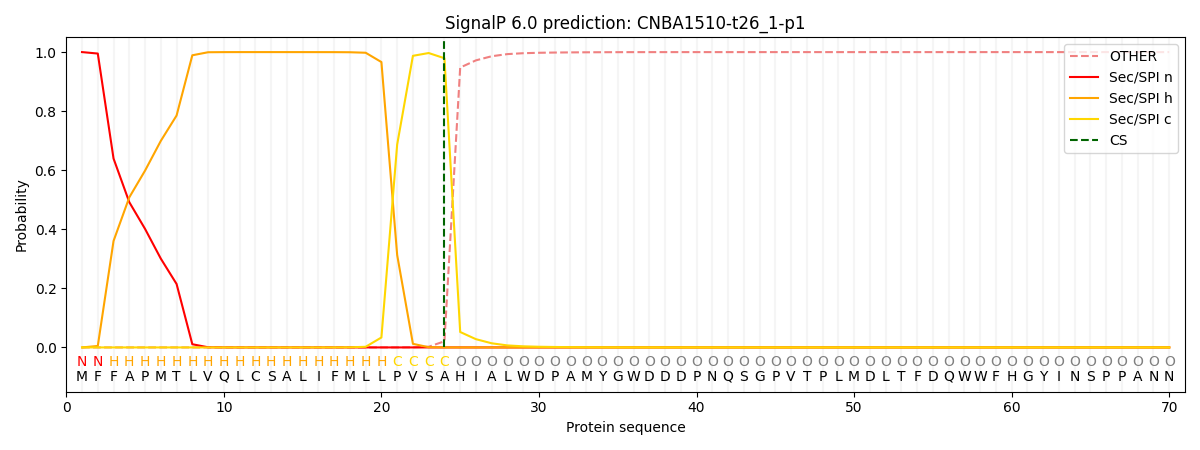
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000329 | 0.999662 | CS pos: 24-25. Pr: 0.9792 |