You are browsing environment: FUNGIDB
CAZyme Information: CLUG_05618-t34_1-p1
You are here: Home > Sequence: CLUG_05618-t34_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
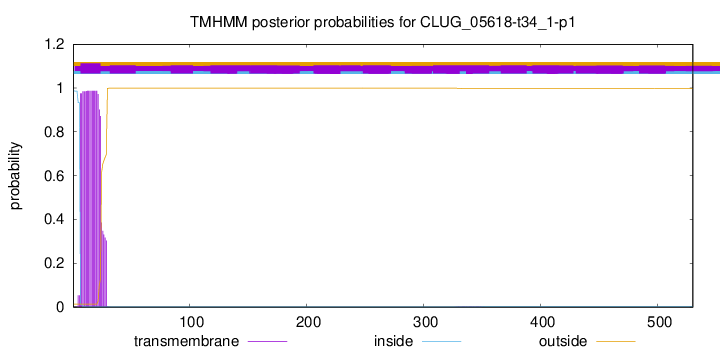
TMHMM annotations
Basic Information help
| Species | Clavispora lusitaniae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Metschnikowiaceae; Clavispora; Clavispora lusitaniae | |||||||||||
| CAZyme ID | CLUG_05618-t34_1-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 148 | 402 | 2e-62 | 0.9962121212121212 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402574 | Mannosyl_trans3 | 2.22e-58 | 148 | 402 | 1 | 273 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 530 | 1 | 530 | |
| 0.0 | 1 | 530 | 1 | 530 | |
| 0.0 | 1 | 530 | 1 | 530 | |
| 0.0 | 1 | 530 | 1 | 530 | |
| 0.0 | 1 | 530 | 1 | 530 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.53e-60 | 137 | 519 | 160 | 555 | Alpha-1,3-mannosyltransferase MNT2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT2 PE=1 SV=1 |
|
| 3.08e-54 | 138 | 519 | 184 | 577 | Probable alpha-1,3-mannosyltransferase MNT4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT4 PE=3 SV=1 |
|
| 2.59e-49 | 84 | 518 | 182 | 726 | Putative alpha-1,3-mannosyltransferase MNN15 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN15 PE=2 SV=2 |
|
| 4.19e-49 | 18 | 518 | 132 | 678 | Putative alpha-1,3-mannosyltransferase MNN14 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN14 PE=2 SV=1 |
|
| 3.07e-43 | 137 | 520 | 387 | 826 | Putative alpha-1,3-mannosyltransferase MNN12 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN12 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
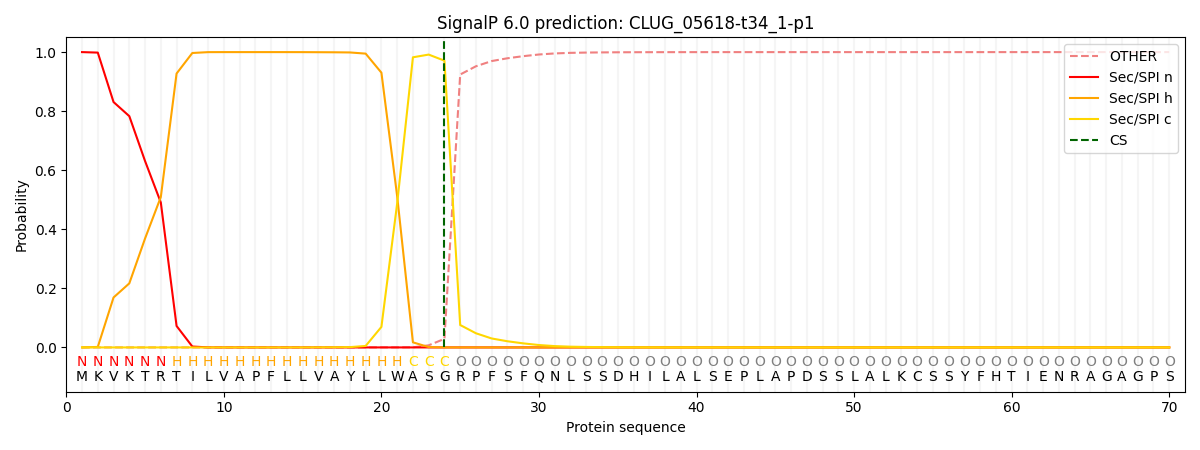
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000294 | 0.999674 | CS pos: 24-25. Pr: 0.9710 |